分子系統学演習
|
|
|
- ぜんま こうだ
- 9 years ago
- Views:
Transcription
1
2
3 i Windows MacOS X Linux EMBOSS Windows MacOS X Linux ClustalW2/X Windows MacOS X Linux MAFFT Windows MacOS X Linux Unipro UGENE Windows MacOS X Linux Kakusan Windows MacOS X Linux Aminosan
4 ii Windows MacOS X Linux Treefinder Windows MacOS X Linux PAUP* Windows MacOS X Linux TNT Windows MacOS X Linux Phylogears Windows MacOS X Linux CONSEL Windows MacOS X Linux trimal Windows MacOS X Linux MrBayes5D Windows MacOS X Linux Tracer Windows MacOS X Linux FigTree
5 iii GenBank FASTA Clustal PHYLIP NEXUS seqret ClustalW2/X Phylogears Treefinder GenBank OTU Mixed model Empirical model Mixed empirical model Mixed model
6 iv 3.2 Kakusan4 Aminosan Treefinder Treefinder Phylogears2 likelihood ratchet Likelihood ratchet Treefinder Treefinder Phylogears MrBayes5D Tracer MrBayes5D MPI Phylogears Treefinder Treefinder Treefinder Treefinder KH SH AU MrBayes5D Bayes factor
7 v UNIX
8
9 ( ) ( ) Second Street, Suite 300, San Francisco, California 94105, USA
10
11 3 0 Treefinder Tracer FigTree Java Windows Java Java MacOS X Java Java Linux Java Java Phylogears2 Perl Windows ActivePerl Windows Perl ActivePerl 32bit C:\Perl MacOS X Linux Perl Windows C:Y=Perl C:\Perl Windows ContextConsole Shell Extension
12 4 0 Windows (.fas.nex ) (Win E ) (Vista ) OK Windows Vista/7 Symantec TrendMicro MacOS X Xcode Tools OS Xcode Mac DVD MacPorts UNIX MacPorts MacPortsWiki-JP MacPorts Fink Homebrew MacPorts
13 5 Fink MacPorts Homebrew MacOS X MacOS X iantivirus OS ( ) Windows XP OS 1 Windows CPU 1 Windows MacOS X Linux nice nice ( 19) ( 19) renice nice OS
14 /10/22 Perl Windows Windows sakura.exe QuickStartV2.zip bregonig.dll Unicode bregonig.dll MacOS X MacOS X CotEditor mi TextWrangler Linux Linux Emacs vim GUI gedit (GNOME ) Kate (KDE )
15 0.2 EMBOSS EMBOSS EMBOSS Windows Windows EMBOSS ftp://emboss.open-bio.org/pub/emboss/windows/ MacOS X Linux MacOS X MacPorts Fink Linux /temp (x.x.x ) cd /temp tar xzf./emboss-x.x.x.tar.gz cd./emboss-x.x.x./configure --prefix=/usr/local --without-x --without-java --without-pngdriver make sudo make install make clean 0.3 ClustalW2/X2 ClustalW2/X2 (multiple sequence alignment)
16 8 0 ClustalW2 ClustalX2 ClustalX Windows ClustalW2 clustalw2.exe C:\Perl\bin MacOS X ClustalX2 (/Applications) Dock ClustalW2 /temp cd /temp sudo mv./clustalw2 /usr/local/bin/ ClustalX2 MacPorts Fink Linux 0.4 MAFFT MAFFT (multiple sequence alignment)
17 0.5 Unipro UGENE Windows All-in-one version mafft.bat ms C:\Perl\bin MacOS X MacOS X Linux /temp cd /temp/mafft-*/core make sudo make install make clean 0.5 Unipro UGENE Unipro UGENE (multiple sequence alignment) Windows MacOS X Linux
18 Windows Windows MAFFT Unipro UGENE MAFFT Settings Preferences... External Tools MAFFT... C:\Perl\bin\mafft.bat clustalw2.exe ClustalW MacOS X MacOS X ugeneui (/Applications) Dock Windows MAFFT ClustalW2 MacOS X Preferences... ugeneui Linux Ubuntu Fedora Windows MAFFT ClustalW2 0.6 Kakusan4 Kakusan Windows Windows Windows Windows kakusan4-4.x.yyyy.mm.dd for Windows.exe
19 0.6 Kakusan4 11 (x yyyy.mm.dd ) Kakusan4 Ctrl+C MacOS X MacOS X kakusan4-4.x.yyyy.mm.dd for MacOSX.zip (x yyyy.mm.dd ) Kakusan4 (/Applications) Dock Kakusan4 Kakusan4 Ctrl+C Linux Linux kakusan4-4.x.yyyy.mm.dd.zip (x yyyy.mm.dd ) baseml Makefile.* baseml Kakusan4 kakusan4.pl ReadSeq PHYLIP ReadSeq ftp://ftp.bio.indiana.edu/molbio/readseq/java/readseq.jar Java readseq.jar Kakusan4 kakusan4.pl ( Java ) PHYLIP Statistics::Distributions Statistics::ChisqIndep 2 Perl Perl CPAN Ctrl+C
20 12 0 kakusan4.pl --interactive=enable 0.7 Aminosan Aminosan Windows Windows Windows Windows aminosan-x.x.yyyy.mm.dd for Windows.exe (x.x yyyy.mm.dd ) Aminosan Ctrl+C MacOS X MacOS X aminosan-x.x.yyyy.mm.dd for MacOSX.zip (x.x yyyy.mm.dd ) Aminosan (/Applications) Dock Aminosan Aminosan Ctrl+C Linux Linux aminosan-x.x.yyyy.mm.dd.zip (x.x yyyy.mm.dd )
21 0.8 Treefinder 13 ReadSeq PHYLIP ReadSeq ftp://ftp.bio.indiana.edu/molbio/readseq/java/readseq.jar Java readseq.jar Aminosan aminosan.pl ( Java ) PHYLIP Statistics::Distributions Statistics::ChisqIndep 2 Perl Perl CPAN Ctrl+C aminosan.pl --interactive=enable 0.8 Treefinder Treefinder Windows Linux Windows Windows Treefinder tf-*-windows.exe (* ) URL tf-march2011-windows.exe URL Treefinder tf.exe C:\Perl\bin Microsoft Visual C SP1 Microsoft MacOS X MacOS X tf-*-mac.zip (* )
22 14 0 tf-october2008-mac.zip URL Treefinder (/Applications) PowerPC G5 Intel CPU tf-october2008-mac-g5-binary.zip tf-october2008-mac-intel-binary.zip tf.bin /Applications/Treefinder/tf.bin Intel CPU Mac cd /Applications chmod -R 755./Treefinder sudo mkdir -p /usr/local/bin sudo ln -f /Applications/Treefinder/tf /usr/local/bin/tf sudo ln -f /Applications/Treefinder/treefinder /usr/local/bin/treefinder treefinder Linux Linux tf-*-linux.zip (* ) Treefinder chmod -R 755./Treefinder sudo mkdir -p /opt sudo mv./treefinder /opt/ sudo ln -s /opt/treefinder/tf /usr/local/bin/tf sudo ln -s /opt/treefinder/treefinder /usr/local/bin/treefinder tf-march2011-linux.zip URL X treefinder
23 0.9 PAUP* PAUP* PAUP* Windows PAUP* win-paup4b10-console.exe paup.exe win-paup4b10-console.exe paup.exe paup.exe C:\Perl\bin Quit paup MacOS X Linux UNIX paup4b10-* paup /temp cd /temp/paup4b10-* chmod 755./paup4b10-* sudo mkdir -p /usr/local/bin sudo mv./paup4b10-* /usr/local/bin/paup PAUP* Quit paup 0.10 TNT TNT
24 16 0 TNT Windows Win (charmode) zipchtnt.zip tnt.exe C:\Perl\bin tnt MacOS X Linux tnt.command (MacOS X ) tnt (Linux ) MacOS X /temp Linux cd /temp chmod 755./tnt.command./tnt.command sudo mv./tnt.command /usr/local/bin/tnt y I agree ( ) TNT tnt TNT
25 0.11 Phylogears Phylogears2 Phylogears2 Perl Treefinder Windows Windows ZIP Windwows phylogears-x.x.yyyy.mm.dd for Windows.zip (x.x yyyy.mm.dd ) ZIP phylogears-x.x.yyyy.mm.dd for Windows bin C:\Perl\bin Perl Math::Random::MT::Perl Math::Random::MT::Auto ( Math::Random::MT::Auto ) Math::Random::MT::Auto Math::Random::MT::Perl ppm install Math-Random-MT-Auto Perl perl -e "use Math::Random::MT::Auto" χ 2 pgtestcomposition Statistics::ChisqIndep Windows ppm install Statistics-ChisqIndep
26 18 0 perl -e "use Statistics::ChisqIndep" MacOS X Linux phylogears-x.x.yyyy.mm.dd.zip (x.x yyyy.mm.dd ) /temp cd /temp/phylogears-x.x.yyyy.mm.dd/bin chmod 755./* sudo mkdir -p /usr/local/bin sudo mv./* /usr/local/bin cd../share sudo mv./phylogears /usr/local/share/ MacOS X Linux Perl Math::Random::MT::Auto sudo -H cpan -i Math::Random::MT::Auto cpan perl -e "use Math::Random::MT::Auto" χ 2 pgtestcomposition Statistics::ChisqIndep sudo -H cpan -i Statistics::ChisqIndep 0.12 CONSEL CONSEL (resampling of estimated log-likelihoods, RELL) Treefinder Treefinder
27 0.13 trimal 19 URL shimo/prog/consel/ Windows consel-0.2 i686-bin C:\Perl\bin 64bit Windows x86 64-bin MacOS X Linux /temp cd /temp tar xzf./cnsls*.tgz cd./consel/src make make install clean cd../bin sudo mv * /usr/local/bin/ 0.13 trimal trimal Web Windows Windows bin C:\Perl\bin
28 MacOS X Linux /temp cd /temp tar xzf./trimal.*.tar.gz cd./trimal/source make sudo mv trimal /usr/local/bin/ sudo mv readal /usr/local/bin/ make clean 0.14 MrBayes5D MrBayes5D MrBayes Windows mrbayes5d.exe C:\Perl\bin mrbayes5d MacOS X mrbayes5d osx.command MacOS X Linux
29 0.15 Tracer Linux /temp (x.x.x yyyy.mm.dd ) cd /temp unzip./mrbayes5d-x.x.x.yyyy.mm.dd.zip cd./mrbayes5d-x.x.x.yyyy.mm.dd make sudo mkdir -p /usr/local/bin sudo mv./mrbayes5d /usr/local/bin/mrbayes5d make clean MPI LAM/MPI OpenMPI MPI MPI cd /temp unzip./mrbayes5d-x.x.x.yyyy.mm.dd.zip cd./mrbayes5d-x.x.x.yyyy.mm.dd MPI=yes make mv./mrbayes5d /mrbayes5d-mpi make clean / mrbayes5d-mpi mpirun -np CPU /mrbayes5d-mpi MPI LAM/MPI mpirun lamboot -v lamhalt OpenMPI 0.15 Tracer Tracer MrBayes MrBayes5D
30 Windows Windows Tracer v*.exe (* ) (C:\Program Files ) MacOS X MacOS X.dmg (/Applications) Dock Linux JAR /temp (* ) cd /temp tar xzf Tracer_v*.tgz cd./tracer_v*/bin chmod 755./tracer sudo mkdir -p /usr/local/bin sudo mv./tracer /usr/local/bin cd../lib sudo mkdir -p /usr/local/lib sudo mv./tracer.jar /usr/local/lib tracer 0.16 FigTree FigTree Tracer
31 GenBank Web (annotation) 1.1 GenBank 1 LOCUS ABC bp 2 DEFINITION TaxonA 18S small subunit ribosomal RNA gene, partial sequence. 3 ORIGIN 4 1 AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 5 // 6 7 LOCUS ABC bp 8 DEFINITION TaxonB 18S small subunit ribosomal RNA gene, partial sequence. 9 ORIGIN 10 1 AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 11 // LOCUS ABC bp 14 DEFINITION TaxonC 18S small subunit ribosomal RNA gene, partial sequence. 15 ORIGIN 16 1 AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 17 //
32 24 1 FASTA Web (annotation) (assemble) (multiple sequence editor) ClustalW/X?? N FASTA 1.2 FASTA 1 >TaxonA 2 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 3 >TaxonB 4 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 5 >TaxonC 6 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA Clustal ClustalW/X (multiple sequence alignment) 1.3 Clustal 1 CLUSTAL multiple sequence alignment TaxonA AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 5 TaxonB AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 6 TaxonC AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 7 ************************************************************ PHYLIP
33 PHYLIP interleaved PHYLIP interleaved 1 1 GenBank Clustal FASTA non-interleaved interleaved non-interleaved PHYLIP 1.4 non-interleaved PHYLIP TaxonA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 3 AAAAAAAAAA 4 TaxonB AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 5 AAAAAAAAAA 6 TaxonC AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 7 AAAAAAAAAA interleaved PHYLIP 1.5 interleaved PHYLIP TaxonA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 3 TaxonB AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 4 TaxonC AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 5 6 AAAAAAAAAA 7 AAAAAAAAAA 8 AAAAAAAAAA 50 non-interleaved interleaved interleaved non-interleaved NEXUS Data interleaved PHYLIP Data
34 GenBank Clustal FASTA 1.6 NEXUS 1 #NEXUS 2 3 Begin Data; 4 Dimensions NTax=3 NChar=60; 5 Format DataType=DNA Interleave Missing=? Gap=-; 6 Matrix 7 TaxonA AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 8 TaxonB AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 9 TaxonC AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA TaxonA AAAAAAAAAA 12 TaxonB AAAAAAAAAA 13 TaxonC AAAAAAAAAA 14 ; 15 End; seqret seqret EMBOSS PHYLIP/NEXUS seqret input_file phylip::output_file seqret input_file nexus::output_file seqret fasta::input_file phylip::output_file
35 ClustalW2/X2 ClustalW2/X2 FASTA EMBL Clustal GCG PHYLIP NEXUS ClustalX2 File Load sequences File Save sequence as... OK Save Sequence As: (File extension will be appended) PHYLIP.phy NEXUS.nxs ClustalW2 PHYLIP/NEXUS clustalw2 -convert -output=phylip -outfile=output_file -infile=input_file clustalw2 -convert -output=nexus -outfile=output_file -infile=input_file Windows -convert /convert - / clustalw2 -options Phylogears2 Phylogears2 FASTA NEXUS PHYLIP Treefinder 4 pgconvseq NEXUS PHYLIP 1 NEXUS PHYLIP FASTA Treefinder FASTA Treefinder % end of data (Phylogears2 ) FASTA NEXUS PHYLIP pgconvseq --output=phylip input_file output_file pgconvseq --output=nexus input_file output_file pgconvseq --output=tf input_file output_file PHYLIP 10 PHYLIPex
36 PHYML RAxML PAML OTU Treefinder Treefinder PHYLIP NEXUS FASTA Utilities Transform Sequence Data... Sequence File Save As OK tf tf TL> SaveSequences[LoadSequences["input_file"],"output_file",Format->"FASTA"] TL> "input_file",loadsequences,"output_file",format->"fasta",savesequences Quit tf command_file
37 NCBI Taxonomy URL NCBI NCBI Taxonomy Nucleotide Protein NCBI Gene URL Nucleotide Protein NCBI Nucleotide Protein [ ] URL , :1000[Sequence Length]
38 30 1 Display GenBank GenBank Show 1 (Sorted By) Send to Text File GenBank Send to File GenBank NCBI BLAST URL BLAST TV URL GenBank GenBank (annotation) GenBank 1.7 D. melanogaster 1 LOCUS NC_ bp DNA circular INV 06-MAY DEFINITION Drosophila melanogaster mitochondrion, complete genome. 3 ACCESSION NC_ VERSION NC_ GI: DBLINK Project :164 6 KEYWORDS. 7 SOURCE mitochondrion Drosophila melanogaster (fruit fly) 8 ORGANISM Drosophila melanogaster 9 Eukaryota; Metazoa; Arthropoda; Hexapoda; Insecta; Pterygota; 10 Neoptera; Endopterygota; Diptera; Brachycera; Muscomorpha; 11 Ephydroidea; Drosophilidae; Drosophila; Sophophora. 12 REFERENCE 1 (bases 1 to 408; to 19517) 13 AUTHORS Lewis,D.L., Farr,C.L. and Kaguni,L.S. 14 TITLE Drosophila melanogaster mitochondrial DNA: completion of the 15 nucleotide sequence and evolutionary comparisons 16 JOURNAL Insect Mol. Biol. 4 (4), (1995) 17 PUBMED FEATURES Location/ Qualifiers 20 source
39 1.3 GenBank /organism=" Drosophila melanogaster" 22 / organelle=" mitochondrion" 23 /mol_type="genomic DNA" 24 /db_xref="taxon:7227" 25 gene /gene="trni" 27 / nomenclature="official Symbol: mt:trna:i Name: 28 mitochondrial isoleucine trna Provided by: FBgn " 29 /note="trna[ile]" 30 /db_xref="flybase: FBgn " 31 /db_xref="geneid :261011" 32 trna /gene="trni" 34 /product="trna -Ile" 35 /db_xref="flybase: FBgn " 36 /db_xref="geneid :261011" gene /gene="nd2" 40 / nomenclature="official Symbol: mt:nd2 Name: 41 mitochondrial NADH - ubiquinone oxidoreductase chain 2 42 Provided by: FBgn " 43 /note="urf2" 44 /db_xref="flybase: FBgn " 45 /db_xref="geneid :192474" 46 CDS /gene="nd2" 48 /note="taa stop codon is completed by the addition of 3 A 49 residues to the mrna" 50 / codon_start=1 51 / transl_except=( pos:1263, aa:term) 52 / transl_table=5 53 /product="nadh dehydrogenase subunit 2" 54 / protein_id=" NP_ " 55 /db_xref="gi: " 56 /db_xref="flybase: FBgn " 57 /db_xref="geneid :192474" 58 / translation=" MFNNSSKILFITIMIIGTLITVTSNSWLGAWMGLEINLLSFIPL 59 LSDNNNLMSTEASLKYFLTQVLASTVLLFSSILLMLKNNMNNEINESFTSMIIMSALL 60 LKSGAAPFHFWFPNMMEGLTWMNALMLMTWQKIAPLMLISYLNIKYLLLISVILSVII 61 GAIGGLNQTSLRKLMAFSSINHLGWMLSSLMISESIWLILFFFYSFLSFVLTFMFNIF 62 KLFHLNQLFSWFVNSKILKFTLFMNFLSLGGLPPFLGFLPKWLVIQQLTLCNQYFMLT 63 IMMMSTLITLFFYLRICYSAFMMNYFENNWIMKMNMNSINYNMYMIMTFFSIFGLFLI 64 SLFYFMF" ORIGIN 67 1 aatgaattgc ctgataaaaa ggattacctt gatagggtaa atcatgcagt tttctgcatt // FEATURES ORIGIN FEATURES ORIGIN extractfeat EMBOSS trni extractfeat -type trna -tag gene -value trni input_file output_file trna trni FASTA
40 32 1 ND2 extractfeat -type CDS -tag gene -value ND2 input_file output_file "ND2 NAD2" 16S ribosomal RNA 100bp -before -after extractfeat -type CDS -tag gene -value ND2 -before 100 -after 100 input_file output_file 1.4 (alignment) (homologous) ( Fleissner et al., 2005; Lunter et al., 2005; Redelings and Suchard, 2005, ) (multiple sequence alignment) ClustalW2/X2 (Larkin et al., 2007) MUSCLE (Edgar, 2004) MAFFT (Katoh et al., 2005) MAFFT
41 MAFFT FASTA mafft --auto input_file > output_file --auto MAFFT ( ) MAFFT EMBOSS tranalign mafft --auto input_file > output_file Unipro UGENE ClustalX EMBOSS sixpack sixpack input_file standard -table invertebrate mitochondrial
42 34 1 sixpack -table 5 input_file -table 0. Standard (default) 1. Standard with alternative initiation codons 2. Vertebrate Mitochondrial 3. Yeast Mitochondrial 4. Mold, Protozoan, Coelenterate Mitochondrial and Mycoplasma/Spiroplasma 5. Invertebrate Mitochondrial 6. Ciliate Macronuclear and Dasycladacean 9. Echinoderm Mitochondrial 10. Euplotid Nuclear 11. Bacterial 12. Alternative Yeast Nuclear 13. Ascidian Mitochondrial 14. Flatworm Mitochondrial 15. Blepharisma Macronuclear 16. Chlorophycean Mitochondrial 21. Trematode Mitochondrial 22. Scenedesmus obliquus 23. Thraustochytrium Mitochondrial sixpack FASTA open reading frame (ORF) ORF ( ) sixpack ORF revseq EMBOSS degapseq degapseq input_file output_file EMBOSS transeq standard -table
43 transeq input_file output_file MAFFT mafft --auto input_file > output_file EMBOSS tranalign standard -table tranalign nonaligned_nucleotide_sequences aligned_peptide_sequences output_file (homologous) 1.1 Y locus Z locus Taxon A Y locus Taxon B Y locus Taxon C Z locus (Taxon B Taxon C Taxon A ) (paralogous) Y locus Z locus 1.1 Taxon A - Y locus Taxon B - Y locus Taxon C - Y locus Duplication Taxon A - Z locus Taxon B - Z locus Taxon C - Z locus (orthologous)
44 36 1 OTU BLAST BLAST Ensembl genome browser Ensembl URL ( 1 1 ) 1 (1 ) ( 0 ) ( ) ( ) missing data ( ) ( Boussau and Gouy, 2006; Blanquart and Lartillot, 2006, 2008, ) OTU
45 OTU OTU RY coding (Woese et al., 1991) Dayhoff coding (Hrdy et al., 2004) (Blanquart and Lartillot, 2006, 2008) ( ) rrna/trna loop (Talavera and Castresana, 2007) Gblocks (Castresana, 2000) trimal (Capella-Gutiérrez et al., 2009) BMGE (Criscuolo and Gribaldo, 2010) trimal trimal PHYLIP FASTA NEXUS trimal 2 trimal -gappyout -in input_file -out output_file trimal -strict -in input_file -out output_file trimal -automated1 -in input_file -out output_file trimal Phylogears2 pgtrimal pgtrimal trimal NEXUS
46 38 1 pgtrimal --frame=1 --method=gappyout input_file output_file pgtrimal --frame=1 --method=strict input_file output_file pgtrimal --frame=1 --method=automated1 input_file output_file pgtrimal --frame --frame= frame=2 2 --frame= RI 1.1 N R Y missing data -? M R W S Y K V H D B N A or C (amino) A or G (purine) A or T C or G C or T (pyrimidine) G or T (keto) A or C or G A or C or T A or G or T C or G or T A or C or G or T [] interleaved
47 1.6 OTU 39 ( ) TNT 31 (.) 1.6 OTU OTU ( ) OTU OTU 1 Phylogears2 pgelimdupseq pgelimdupseq --type=dna input_file output_file --type=dna --type=aa 1 (OTU ) 2 FASTA NEXUS PHYLIP extended PHYLIP Treefinder PHYLIP 10 A G R A C G T N A G A R AAA ARA R R AAA R DNA DNA
48 40 1 A G R ARA R R ( ) AAA ARA pgelimdupseq AAA ARA --prefer=degenerate --prefer=both pgelimdupseq pgelimdupseq -? (missing data, - N ) --gap=another 1.7 OTU OTU OTU Kakusan4 Aminosan Phylogears2 pgtestcomposition pgtestcomposition χ 2 PAUP*(Swofford, 2003) BaseFreqs PAUP* pgtestcomposition PAUP* R A G 0.5 pgtestcomposition Bowker (Ababneh et al., 2006) p pgtestcomposition pgtestcomposition --type=dna input_file output_file --type=dna --type=aa FASTA NEXUS PHYLIP extended PHYLIP Treefinder
49 χ 2 1 Type of Nucleotides: 4 2 Number of Taxa: 3 Degree of Freedom: 4 Total Count: * - Gap Missing Data Ambiguous Data 5 Chi -square Statistic: chi - s q u a r e 6 p-value: p 7 8 A C G T rtotal 9 OTU ctotal (Blanquart and Lartillot, 2006, 2008) p (Cochran, 1954) pgtestcomposition --type=dna CG TA input_file output_file 3 pgtestcomposition --type=dna 3-.\3 CG TA input_file output_file 3-.\3 3 3 (2 ) RY (Woese et al., 1991) RY AT GC OTU AG CT OTU A G ( R ) T C ( Y ) 2 AG TC Phylogears2 pgrecodeseq RY
50 42 1 pgrecodeseq --type=dna CG TA input_file output_file C T G A A T 2 ( -? ) RY 2 FASTA NEXUS PHYLIP extended PHYLIP Treefinder Dayhoff (Hrdy et al., 2004) pgrecodeseq --type=aa STGPNEQKHVILYW AAAADDDRRMMMFF input_file output_file ADRMFC 6 RAxML (Stamatakis, 2006) Treefinder (Jobb et al., 2004) MrBayes (Ronquist and Huelsenbeck, 2003) (GTR) WAG (Whelan and Goldman, 2001) JTT (Jones et al., 1992) +F Dayhoff pgrecodeseq pgtestcomposition 3 1 pgtestcomposition pgtestcomposition OTU
51 43 2 (nucleotide substitution model) (amino acid substitution model) (synonymous substitution) (nonsynonymous substitution) (codon substitution model) (nucleotide substitution rate matrix) (site) (character state) (heterogeneity) From To A C G T A - Rate AC Freq C Rate AG Freq G Rate AT Freq T C Rate AC Freq A - Rate CG Freq G Rate CT Freq T G Rate AG Freq A Rate CG Freq C - Rate GT Freq T T Rate AT Freq A Rate CT Freq C Rate GT Freq G -
52 44 2 Rate XY Freq X Y X Freq X X Rate XY = Rate YX ( [time-reversible] ) Rate AC = Rate AG = Rate AT = Rate CG = Rate CT = Rate GT Freq A = Freq C = Freq G = Freq T JC69 (Jukes and Cantor, 1969) Rate AG = Rate CT Rate AC = Rate AT = Rate CG = Rate GT Freq A = Freq C = Freq G = Freq T K80/K2P (Kimura, 1980) Rate AC = Rate AG = Rate AT = Rate CG = Rate CT = Rate GT Freq A Freq C Freq G Freq T F81 (Felsenstein, 1981) Rate AC Rate AG Rate AT Rate CG Rate CT Rate GT Freq A Freq C Freq G Freq T (Tavaré, 1986) (general time-reversible GTR) (Posada and Crandall, 1998) GTR ( ) (site) (heterogeneity) ARSV (among-site rate variation) Γ (Yang, 1993) Γ Γ (Yang, 1994) + G + dg (discrete Gamma ) + dg 4 (invariable site) (variable site) 2 ( + I ) + G + I (partitioning) ( + SS [site specific rate ] ) (codon position) + SS + Codon Position Specific Rate + Gene Specific Rate + G + I ( + I ) + Codon Position Specific Rate + G Γ
53 ( + 3 Different Gamma ) Γ ( + 1 Shared/Common Gamma ) Γ ( + N Different Gamma ) Γ ( + 1 Shared/Common Gamma ) + G + adg (autocorrelated discrete Gamma ) (Yang, 1995) Mixed model (site) (partition) ASRV mixed model (partitioned model) ASRV (nonpartitioned model) Mixed model 2 1 (proportional model) 1 (separate model) -1 = ASRV + SS ASRV Empirical model 4x4 20x20 Rate XY Freq X = 210 Rate XY Freq X empirical model (Dayhoff et al., 1978; Henikoff and Henikoff, 1992; Jones et al., 1992; Müller and Vingron, 2000; Whelan and Goldman, 2001; Veerassamy et al., 2003; Le and Gascuel, 2008) (Adachi and Hasegawa, 1996; Cao et al., 1998; Abascal et al., 2007) (Adachi et al., 2000) (Dimmic et al., 2002; Nickle et al., 2007) Rate XY
54 46 2 empirical model Freq X + F Mixed empirical model Empirical model empirical model 20x20 empirical model (Jobb, 2008) Treefinder MrBayes (Ronquist et al., 2005) MrBayes empirical model model jumping (model averaging) Mixed model mixed model
55 Akaike (1974) (Akaike information criterion AIC) AIC L k AIC = 2 ln L + 2k (3.1) AIC AIC AIC AICc Sugiura (1978) AICc n AICc = 2 ln L + 2k n n k 1 (3.2) AICc n k 1 0 AICc
56 48 3 BIC (Schwarz, 1978) BIC = 2 ln L + k ln n (3.3) AIC AICc BIC AIC AICc AICc n k 1 > 0 AICc AIC 1 ( ) model jumping 3.2 Kakusan4 Aminosan Kakusan4 (Tanabe, 2007) Aminosan Treefinder MrBayes (MrBayes5D) PAUP* baseml Treefinder (Aminosan Treefinder codeml) CPU CPU FASTA NEXUS PHYLIP GenBank AIC (Akaike, 1974) AICc (Sugiura, 1978) BIC (Schwarz, 1978) Kakusan4 Aminosan
57 3.2 Kakusan4 Aminosan χ 2 2. ( ) JC69 (Kakusan4) K83 (Aminosan) Kakusan4 Aminosan 2 1 ( ) 2 Kakusan4 Aminosan Aminosan mixed empirical model Kakusan4 Aminosan Kakusan ======================================================================= This is a script to select nucleotide substitution model for multipartitioned data set. Official web site of this script is To know script details, see above URL. Copyright (C) Akifumi S. Tanabe This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License. This program is distributed in the hope that it will be useful,
58 50 3 but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA USA. Parsing command line options... No input files are specified. Entering interactive mode. Specified options are ignored. Specify an input file name. Note that you can use wild card. Windows (Vista ) MacOS X 1 Windows Vista Shift Specify an input file name. Note that you can use wild card. "C:\Users\akifumi\Desktop\SampleData\CYTBnuc_P.fas" Enter Kakusan4 Aminosan "C:\Users\akifumi\Desktop\SampleData\CYTBnuc_P.fas" "C:\Users\akifumi\Desktop\SampleData\CYTBnuc_P.fas" was accepted. Specify an input file name or just press enter to leave input file specification. 5 3 ( ) P mixed model *? Aminosan mt mtrev (Adachi and Hasegawa, 1996) mtmam (Cao et al., 1998) mtart (Abascal et al., 2007) mtzoa (Rota-Stabelli et al., 2009) nc Dayhoff (Dayhoff et al., 1978) JTT (Jones et al., 1992) BLOSUM62 (Henikoff and Henikoff, 1992) VT (Müller and Vingron, 2000) WAG (Whelan and Goldman, 2001) PMB (Veerassamy et al., 2003) LG (Le and Gascuel, 2008) cp cprev (Adachi et al., 2000) rt rtrev
59 3.2 Kakusan4 Aminosan 51 (Dimmic et al., 2002) HIVb HIVw (Nickle et al., 2007) + F, + G, + I Aminosan Dayhoff JTT Kosiol and Goldman (2005) DCMut Enter Specify an input file name or just press enter to leave input file specification. OK. Input file specification have terminated. Log, result and configuration files will be output to "C:\Users\akifumi\Desktop\ SampleData\CYTBnuc_P.fas.kakusan"..kakusan (Aminosan aminosan ) OUTPUT OPTIONS Which is a target analysis software? (MrBayes/Treefinder/PAUP/PHYML/RAxML) (default: Treefinder) Treefinder RAxML MrBayes mixed model RAxML PAUP* PHYML PAUP* PHYML mixed model mixed model Aminosan ANALYSIS OPTIONS You input protein coding sequence. Do you want to consider partitioning of codon positions? (y/n) (default: n) y Enter
60 52 3 PAUP* PHYML Aminosan You enabled partitioning of codon positions. Do you want to consider nonpartitioning of codon positions? (y/n) If you say yes, applying nonpartitioned models to all-codon position-concatenate d sequences will be considered on each locus. (default: n) n Enter y PAUP* PHYML You input multiple files. Do you want to consider nonpartitioning of loci? (y/n) If you say yes, applying nonpartitioned models to all-loci-concatenated sequence s will be considered. (default: n) y Enter n PAUP* PHYML RAxML You input multiple files or protein coding sequence. Do you want to compare nonpartitioned, proportional and separate models on allloci concatenated sequences? (y/n) Note that this function needs Treefinder. (default: y) y Enter Treefinder Treefinder Treefinder Treefinder
61 3.2 Kakusan4 Aminosan 53 Treefinder + SS + SS PAUP* baseml (Aminosan codeml) Treefinder Which do you want to use the program for likelihood calculation? (baseml/tf/paup) (default: baseml) baseml baseml tf Treefinder paup PAUP* Treefinder MrBayes Treefinder baseml (codeml) PAUP* PHYML PAUP* MrBayes Treefinder Treefinder RAxML Do you want to optimize the parameters of base composition? (y/n) (default: n) n Enter y 20 Treefinder Treefinder MrBayes Γ How many rate categories of discrete gamma rate heterogeneity do you want to con sider? (integer) (default: 8) 4 ASRV + I PAUP* Treefinder
62 54 3 Do you want to consider invariant model for among-site rate variation? (y/n) (default: n) n y Γ baseml Do you want to consider N-GAM model for among-site rate variation? (y/n) Note that this model is very time-consuming. (default: n) y Enter Γ n Γ Γ baseml Do you want to consider autocorrelated discrete gamma model for among-site rate variation? (y/n) Note that this model is very time-consuming. (default: n) y Enter Γ Do you want to use different tree topology for parameter optimization on each lo cus? (y/n) (default: n) y Enter n Enter (incongruence) y JC69 (Aminosan K83 [Kimura, 1983])
63 3.2 Kakusan4 Aminosan 55 (neighbor-joining [Saitou and Nei, 1987]) If you want to give tree(s) for parameter optimization, specify an input file na me. Otherwise, just press enter. Newick NEXUS Enter How many processes do you want to run simultaneously? (integer) (default: 1) Enter PC CPU( ) PC All configurations have been completed. Just press enter to run! Enter kakusan (Aminosan aminosan) ( ) Chisq Results MrBayes PAUP PHYML RAxML Treefinder Scores Logs Chisq chisq_partition.txt ( )... Results partition_criterion.txt ( ) whole_criterion_comparemix.txt ( )
64 MrBayes partition_criterion_xxx.nex ( NEXUS )... PAUP partition_criterion.nex ( NEXUS )... PHYML partition.phy ( ) partition_criterion_singlesearch.bat ( ) partition_criterion_shotgunsearch.bat ( ) partition_criterion_bootstrap.bat ( ) partition_criterion_shotgunbootstrap.bat ( )... RAxML partition.phy ( ) partition_criterion_xxx.partition ( ) partition_criterion_xxx_singlesearch.bat ( ) partition_criterion_xxx_shotgunsearch.bat ( ) partition_criterion_xxx_bootstrap.bat ( )... Treefinder partition_xxx.tf ( ) partition_criterion_xxx.model ( ) partition_criterion_xxx.rates ( ) partition_criterion_comparemodels.tl ( Treefinder Language ) partition_criterion_xxx_singlesearch.tl ( Treefinder Language ) partition_criterion_xxx_shotgunsearch.tl ( Treefinder Language ) partition_criterion_xxx_bootstrap.tl ( Treefinder Language )... Scores partition_model.txt ( )... Logs ( )... partition ( ) criterion xxx whole Windows χ 2 (chisq partition.txt) pgtestcomposition p 0.05 OTU p (Blanquart and Lartillot, 2006, 2008) nhphylobayes (partition criterion.txt) model criterion weight -LnL nparam 2 SYM_GeneCodonPos1Gamma e e J2ef_GeneCodonPos1Gamma e e SYM_Gamma e e criterion weight - L n L
65 3.2 Kakusan4 Aminosan 57 GeneCodonPos1Gamma Γ AICc BIC AICc BIC AICc1 BIC1: ( ) AICc2 BIC2: AICc3 BIC3: AICc4 BIC4: ( ) AICc5 BIC5: AICc6 BIC6: AICc4 BIC4 Results whole criterion comparemix.txt model AIC -LnL nparam 2 Separate_CodonProportional e e Proportional_CodonProportional e e Separate_CodonSeparate e e Proportional_CodonNonpartitioned e e Separate_CodonNonpartitioned e e Nonpartitioned e e criterion - L n L
66 58 3 Kakusan4 Aminosan MrBayes (MrBayes5D) Treefinder Kakusan4 Aminosan AIC AICc BIC Kakusan4 Aminosan Treefinder Treefinder 3.3 Kakusan4 Aminosan Treefinder partition criterion comparemodels.tl Treefinder Kakusan4 Aminosan Kakusan4 partition criterion comparemodels.tl partition criterion comparemodels.log 3.3 partition criterion comparemodels.log 1 { 2 {Likelihood ->, Phylogeny ->, SubstitutionModel ->, OSubstitutionModel -> ( ), OEdgeOptimizationOff ->, NSites -> ( ), NParameters ->, AIC -> AIC,AICc -> AICc,HQ-> HQ,BIC -> BIC,Checksum ->, PartitionKeys -> ID,PartitionRates ->, OPartitionRates ->, NSitesPartitionwise ->
67 3.3 59, FilterNames ->, LikelihoodTime ->, LikelihoodMemory -> }, 3 4 () 5 } Treefinder AICc BIC Kakusan4 Aminosan AICc4 BIC4 OPartitionRates-> PartitionKeys-> OPartitionRates->Optimum OPartitionRates->{1:1.,2:1.,3:1.,4:1., } PartitionKeys-> PartitionKeys-> OPartitionRates->{1:1.,2:Optimum,2:Optimum,2:Optimum,3:1., } :Optimum :1.
68
69 :9 L 1 ) 9 L 1 = 1 ( = (4.1) L 0 ( ) 10 1 L 0 = 2 = (4.2) L 1 > L 0 1:9 1 AIC { ( ) ( ) } 1 9 AIC 1 = 2 ln + ln = 8.50 (4.3) { ( ) } 1 AIC 0 = 2 ln = (4.4) AIC 1 < AIC 0
70 62 4 ( ) = = =OTU (operational taxonomic unit) (exhaustive search) (heuristic search) (neighbor-joining [Saitou and Nei, 1987]) (stepwise/sequential sequence addition [Swofford and Begle, 1993]) (initial/starting tree) (branch swapping) (topology rearrangement) 4.2 Treefinder Treefinder (Jobb et al., 2004) RAxML (Stamatakis, 2006) Treefinder RAxML (100 OTU 10k bp 5k aa ) RAxML RAxML GTR Treefinder RAxML Treefinder RAxML RAxML ver h Kakusan4 Aminosan Treefinder partition criterion xxx singlesearch.tl partition criterion xxx mixed model whole whole AIC proportional codonproportional singlesearch.tl (AIC Treefinder Language ) whole AIC codonproportional singlesearch.tl
71 4.2 Treefinder 63 (AIC Treefinder Language ) whole AIC nonpartitioned singlesearch.tl (AIC Treefinder Language ) whole AIC nonpartitioned singlesearch.tl whole AIC codonnonpartitioned singlesearch.tl Kakusan4 p Treefinder Kernel Load TL Script... (*.tl ) tf *.tl ( optimum.model optimum.rates ) ( optimum.nwk).log (TL Report ) optimum.nwk.log File Open Image... Treefinder View Redraw... File Save PostScript 1 Treefinder partition criterion xxx shotgunsearch.tl
72 64 4 (OTU 3) / 2 (nearest neighbor interchange) OTU 3 likelihood ratchet 4.3 Treefinder Phylogears2 likelihood ratchet Ratchet (Nixon, 1999; Vos, 2003) ( ) ratchet ( ) ratchet (parsimony criterion) (random sequence addition [Swofford and Begle, 1993]) likelihood ratchet Vos (2003) PAUP* POY4 TNT
73 4.3 Treefinder Phylogears2 likelihood ratchet 65 ratchet Kakusan4 Aminosan Treefinder Phylogears2 pgtfratchet pgtfratchet pgtfratchet ======================================================================= Official web site of this script is To know script details, see above URL. Copyright (C) Akifumi S. Tanabe This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA USA. Model and TF files were found. Entering interactive mode... Which do you want to analyze? (name/number) 1: CYTBnuc_P 2: ND5nuc_P 3: whole Enter Which model do you want to apply to the data? (name/number) 1: proportional_codonproportional 2: separate_codonproportional 3: separate_codonseparate 4: proportional_codonnonpartitioned 5: separate_codonnonpartitioned 6: nonpartitioned Enter
74 66 4 Which criterion do you want to use? (name/number) 1: AIC 2: AICc1 3: AICc2 4: AICc3 5: AICc4 6: AICc5 7: AICc6 8: BIC1 9: BIC2 10: BIC3 11: BIC4 12: BIC5 13: BIC6 Enter Which do you want to use the program for generation of starting trees? (paup/poy /tf/tnt) (default: poy) PAUP* POY4 TNT Treefinder TNT PAUP* >> POY4 >> Treefinder TNT OTU 31 How many percentages of sites do you want to upweight? (integer) (default: 25) % 1,000bp 25% % How many replicates do you want to run? (integer) (default: 100) pgtfratchet pgtfratchet If you want to give topological constraint, specify an input file name. Otherwise, just press enter.
75 4.3 Treefinder Phylogears2 likelihood ratchet 67 p Enter 2 2 CPU( ) CPU( ) How many processes do you want to run simultaneously? (integer) (default: 1) All configurations have been completed. Just press enter to run! Enter pgtfratchet or ( optimum.model optimum.rates ) ( optimum.nwk).log (TL Report ) Likelihood ratchet Likelihood ratchet
76 68 4 pgtfratchet checkcoverage.txt 1 pgtfratchet 1 # 0: same topology 2 # 1: different topology 3 4 source input same or not checkcoverage.txt source input same or not (0) (1) 1 20 OTU Treefinder (credibility) (bootstrap resampling) (internal/interior branch) (Felsenstein, 1985) Kakusan4 partition criterion xxx bootstrap.tl Treefinder Kernel Load TL Script ( optimum.model optimum.rates ) ( optimum.nwk)
77 4.4 Treefinder 69 bootstrap.log bootstrap.nwk Newick consensus.nwk Treefinder Phylogears2 Kakusan4 Aminosan Treefinder Phylogears2 pgtfboot pgtfboot pgtfboot ======================================================================= Official web site of this script is To know script details, see above URL. Copyright (C) Akifumi S. Tanabe This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA USA. Model and TF files were found. Entering interactive mode... Which do you want to analyze? (name/number) 1: CYTBnuc_P 2: ND5nuc_P 3: whole pgtfratchet Which model do you want to apply to the data? (name/number) 1: proportional_codonproportional 2: separate_codonproportional 3: separate_codonseparate
78 70 4 4: proportional_codonnonpartitioned 5: separate_codonnonpartitioned 6: nonpartitioned Which criterion do you want to use? (name/number) 1: AIC 2: AICc1 3: AICc2 4: AICc3 5: AICc4 6: AICc5 7: AICc6 8: BIC1 9: BIC2 10: BIC3 11: BIC4 12: BIC5 13: BIC6 How many replicates do you want to run? (integer) (default: 100) pgtfratchet If you want to give topological constraint, specify an input file name. Otherwise, just press enter. p Enter Which do you want to use as starting tree? (RAWML/NJ/FILENAME) (default: RAWML) RAWML ( optimum.nwk ) NJ Which value do you want to use to the parameters? (RAWML/OPTIMIZE/FILENAME) (default: RAWML)
79 4.4 Treefinder 71 RAWML ( optimum.model optimum.rates ) OPTIMIZE How many processes do you want to run simultaneously? (integer) (default: 1) 2 2 CPU( ) CPU( ) All configurations have been completed. Just press enter to run! Enter bootstrap.log bootstrap.nwk Newick consensus.nwk optimum with supportvalues.nwk Newick allhypotheses.nwk Newick FigTree
80
81 73 5 (Markov chain Monte Carlo MCMC) (Bayesian phylogenetic inference) (convergence) MCMC MrBayes (Ronquist and Huelsenbeck, 2003) MrBayes5D Tracer 5.1 MrBayes (MrBayes5D) MCMC (Metropolis-Hastings algorithm [Metropolis et al., 1953; Hastings, 1970]) MCMC % (steady state) (burn-in) (posterior distribution) (posterior probability)
82 MrBayes5D MrBayes5D MrBayes MPI MrBayes Kakusan4 Aminosan MrBayes NEXUS MrBayes partition criterion xxx.nex partition criterion xxx whole whole BIC4 proportional codonproportional.nex ( [ ] BIC NEXUS ) whole BIC4 codonproportional.nex ( [ ] BIC NEXUS ) whole BIC4 nonpartitioned.nex ( [ ] BIC NEXUS ) whole BIC4 nonpartitioned.nex whole BIC4 codonnonpartitioned.nex Kakusan4 Kakusan4 Aminosan
83 5.3 Tracer 75 Treefinder MrBayes5D NEXUS MrBayes5D ( ) RAxML MrBayes5D MCMC mrbayes5d -i partition_criterion_xxx.nex MrBayes > MCMC MCMC (NGen 1,000,000 ) MCMC p Tracer MCMC Tracer MrBayes5D ASDSF ASDSF 1,000 MCMC DiagnFreq=10000 (10,000 ASDSF ) MCMCDiagn=No ASDSF NRuns=1 MCMC 1 ASDSF MrBayes5D MCMC MrBayes5D Tracer MCMC NGen 1,000,000 MrBayes > MCMC Continue with analysis? (yes/no): Tracer File Import Trace File... MrBayes5D NEXUS NEXUS.run1.p NEXUS.run2.p 2 Trace Files 2
84 76 5 Ctrl Shift 2 MrBayes5D 2 ( ) MCMC 2 MCMC Tracer Trace Colour by Trace File Legend None 2 MCMC ( 5.1) Traces (steady state) MCMC MCMC 2 MCMC MCMC 5.1 Tracer 70 2 MCMC
85 5.3 Tracer 77 ( ) 2 MCMC Trace Files Burn-In ( ) 1 1,000,000 burn-in 1,000, (SampleFreq=100) 1, ,001,000 burn-in SampleFreq MrBayes5D (summarize) Burn-In MrBayes5D p Burn-In Trace Files Combined Marginal Density Colour by Trace File Legend None MCMC MCMC ( 5.2) Traces MCMC Traces ESS (effective sample size,, [Kass et al., 1998]) MCMC Estimates Traces MCMC ESS ESS (proposal) (acceptance rate) (state exchange) ESS MCMC
86 Tracer MCMC ESS 100 MCMC 2 MCMC ESS ESS ESS ESS ESS MCMC Acceptance rates for the moves in the "cold" chain: With prob. Chain accepted changes to 1.23 % param. 1 (state frequencies) with Dirichlet proposal ESS Props MCMC Tracer
87 5.3 Tracer 79 Tracer Props MrBayes > Props Select a parameter to change (1-36; 0 to exit; 37 to zero all proposal rates): 26 ( ) Proposal 26: Change (rate multiplier) with Dirichlet proposal New proposal rate (<return> to keep old = 1.000): ( Enter) New Dirichlet parameter (<return> to keep old = ): ( ) Select a parameter to change (1-36; 0 to exit; 37 to zero all proposal rates): 0 ( ) proposal rate ( ) MrBayes MCMC MCMC MCMC MCMC MrBayes5D (rate multiplier) Dirichlet proposal Dirichlet parameter ( ) 1000 ( MrBayes 500) MrBayes5D 2 MCMC 2 MCMC 4 MCMC 4 (temperature) (heated chain) 3 (temperature ) (cold chain) 1 MCMC Metropolis-coupled MCMC MC 3 ESS (state exchange) Metropolis et al.
88 80 5 (1953) Hastings (1970) MCMC Chain swap information for run 1: Upper diagonal: Proportion of successful state exchanges between chains Lower diagonal: Number of attempted state exchanges between chains ( 0.2) MrBayes > MCMCP Temp=0.15 MCMC MCMC MCMC 5.4 MCMC burn-in ( ) Tracer burn-in ( ) 1,000,000 burn-in 10,001 (MrBayes5D 1 1 ) p75 p5.3 MrBayes5D.t MrBayes5D SumT Phylogears2 MCMC burn-in SumT MrBayes5D NEXUS integer burn-in
89 5.5 MrBayes5D MPI 81 MrBayes > SumT BurnIn=integer.con.parts.con MCMC (internal/interior branch).parts Phylogears2 Phylogears2 pgsplicetree pgsplicetree from-to input_file output_file from-to ,002 10,001 burn-in t ( 2 ).t pgjointree pgjointree input_file1 input_file2 output_file 3 pgsumtree pgsumtree p p MrBayes5D MPI MrBayes5D MPI (Altekar et al., 2004) / mrbayes5d-mpi mpirun -np CPU /mrbayes5d-mpi -i NEXUS MPI LAM/MPI mpirun lamboot -v lamhalt
90 82 5 Props mcmc.c SetUpMoveTypes MrBayes5D 4 (NChains) 2 (NRuns) 8 MCMC 8 CPU 1 CPU CPU 1 1 (NSwaps) NRuns CPU
91 (clade) OTU (internal/interior branch) (monophyly) (paraphyly) (polyphyly) (monophyletic group) OTU (paraphyletic group) 6.1 TaxonA TaxonB TaxonC TaxonD OTU OTU 6.1 (TaxonA, TaxonB) (TaxonC,
92 84 6 TaxonD) (TaxonA, TaxonB, TaxonC, TaxonD) (TaxonA, TaxonB, TaxonC) (TaxonA, TaxonB, TaxonD) (TaxonA, TaxonC, TaxonD) (TaxonB, TaxonC, TaxonD) OTU (TaxonA, TaxonC) (TaxonA, TaxonD) (TaxonB, TaxonC) (TaxonB, TaxonD) ( ) OTU (ancestral/plesiomorphic) (derived/apomorphic) ( OTU ) ( ) 6.2 PHYLIP/Newick NEXUS PHYLIP/Newick 6.1 PHYLIP/Newick (TaxonA:0.1, TaxonB:0.1,( TaxonC:0.1, TaxonD :0.1):0.1); 3 (TaxonA:0.1, TaxonC:0.1,( TaxonB:0.1, TaxonD :0.1):0.1); 4 (TaxonA:0.1, TaxonD:0.1,( TaxonB:0.1, TaxonC :0.1):0.1); (:) PHYLIP OTU 10 Newick NEXUS 6.2 NEXUS 1 #NEXUS 2 3 Begin Trees; 4 tree tree_1 = [&U] (TaxonA:0.1, TaxonB:0.1,( TaxonC:0.1, TaxonD :0.1):0.1); 5 tree tree_2 = [&U] (TaxonA:0.1, TaxonC:0.1,( TaxonB:0.1, TaxonD :0.1):0.1); 6 tree tree_3 = [&U] (TaxonA:0.1, TaxonD:0.1,( TaxonB:0.1, TaxonC :0.1):0.1); 7 End; Trees [&U] [&R] Translate OTU
93 Translate NEXUS 1 #NEXUS 2 3 Begin Trees; 4 Translate 5 1 TaxonA, 6 2 TaxonB, 7 3 TaxonC, 8 4 TaxonD 9 ; 10 tree tree_1 = [&U] (1:0.1,2:0.1,(3:0.1,4:0.1):0.1); 11 tree tree_2 = [&U] (1:0.1,3:0.1,(2:0.1,4:0.1):0.1); 12 tree tree_3 = [&U] (1:0.1,4:0.1,(2:0.1,3:0.1):0.1); 13 End; Phylogears2 Phylogears2 pgconvtree PHYLIP/Newick NEXUS Treefinder TL Report Newick/PHYLIP NEXUS pgconvtree --output=newick input_file output_file pgconvtree --output=nexus input_file output_file Translate NEXUS Treefinder Treefinder PHYLIP/Newick NEXUS Phylogears2 Translate NEXUS File Open Image... File Export Tree... Save As OK tf tf
94 86 6 TL> SaveTreeList[LoadTreeList["input_file"],"output_file",Format->"NEWICK"] TL> "input_file",loadtreelist,"output_file",format->"newick",savetreelist Quit tf command_file Treefinder Treefinder 1 Mesquite Mesquite File Open Image... ( 6.2) View Redraw Treefinder 6.3 Reroot
95 OTU 2 OTU 2 OTU 6.3 TaxonD TaxonE OK Treefinder Swap Group 1 Group 2 OTU 2 OTU 2 OTU 6.4 Group 1 TaxonE TaxonF Group 2 TaxonA OK Treefinder OTU (Remove Tips) 0 (Collapse) tf tf
96 88 6 ( ) TL> SaveTreeList[RedrawTree[LoadTreeList["input_file"],Outgroup->{"TaxonA","TaxonB"}], "output_file",format->"newick"] 1OTU {} TL> "input_file",loadtreelist,outgroup->{"taxona","taxonb"},redrawtree,"output_file", Format->"NEWICK",SaveTreeList OTU Outgroup->{"TaxonA","TaxonB"} GroupsToSwap->{{"TaxonA","TaxonB"},{"TaxonC","TaxonD"}} OTU TipsToRemove->{"TaxonA","TaxonB"} Quit tf command_file a 6.5b, c 6.5b, c ( ) 6.5b-e Phylogears2 pgsumtree MCMC pgtfboot (p bootstrap.nwk ) pgsumtree --mode=all *_bootstrap.nwk output_file
97 a OTU1 OTU2 OTU3 OTU4 OTU5 b 6.5 OTU1 OTU2 OTU3 OTU4 OTU5 d OTU1 OTU3 OTU2 OTU4 OTU5 c OTU1 OTU2 OTU3 OTU4 OTU5 e OTU1 OTU3 OTU5 OTU2 OTU4 a b, c a b, c b-e Newick 16OTU 100 pgsumtree [ majorhypothesis_1] (( TaxonA,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonG,TaxonH,TaxonI, TaxonJ,TaxonK,TaxonL,TaxonM,TaxonN)100.0,( TaxonO,TaxonP)); 2 [ majorhypothesis_2] (( TaxonA,TaxonO,TaxonP,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonG, TaxonH,TaxonI,TaxonJ,TaxonM,TaxonN)100.0,( TaxonK,TaxonL)); 3 [ majorhypothesis_3] (( TaxonA,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonH,TaxonI,TaxonJ, TaxonK,TaxonL,TaxonM)100.0,( TaxonO,TaxonP,TaxonG,TaxonN)); 4 [ majorhypothesis_4] (( TaxonA,TaxonO,TaxonP,TaxonB,TaxonE,TaxonF,TaxonG,TaxonH,TaxonI, TaxonJ,TaxonK,TaxonL,TaxonM,TaxonN)100.0,( TaxonC,TaxonD)); 5 [ majorhypothesis_5] (( TaxonA,TaxonO,TaxonP,TaxonC,TaxonD,TaxonF,TaxonG,TaxonH,TaxonI, TaxonJ,TaxonK,TaxonL,TaxonM,TaxonN)98.0,( TaxonB,TaxonE)); 6 [ majorhypothesis_6] (( TaxonA,TaxonO,TaxonP,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonH, TaxonI,TaxonJ,TaxonK,TaxonL,TaxonM)85.0,( TaxonG,TaxonN)); 7 8 [ minorhypothesis_1] (( TaxonA,TaxonO,TaxonP,TaxonB,TaxonE,TaxonF,TaxonG,TaxonH,TaxonJ, TaxonK,TaxonL,TaxonM,TaxonN)25.0,( TaxonC,TaxonD,TaxonI)); 9 [ minorhypothesis_2] (( TaxonA,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonH,TaxonI,TaxonJ, TaxonK,TaxonL)21.0,( TaxonO,TaxonP,TaxonG,TaxonM,TaxonN)); 10 [ minorhypothesis_3] (( TaxonA,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonH,TaxonI,TaxonK, TaxonL,TaxonM)17.0,( TaxonO,TaxonP,TaxonG,TaxonJ,TaxonN)); 11 [ minorhypothesis_4] (( TaxonA,TaxonH,TaxonJ)15.0,( TaxonO,TaxonP,TaxonB,TaxonC,TaxonD, TaxonE,TaxonF,TaxonG,TaxonI,TaxonK,TaxonL,TaxonM,TaxonN)); 12 [ minorhypothesis_5] (( TaxonA,TaxonO,TaxonP,TaxonB,TaxonE,TaxonF,TaxonG,TaxonH,TaxonJ, TaxonK,TaxonL,TaxonN)14.0,( TaxonC,TaxonD,TaxonI,TaxonM)); 13 [ minorhypothesis_6] (( TaxonA,TaxonC,TaxonD,TaxonM)12.0,( TaxonO,TaxonP,TaxonB,TaxonE, TaxonF,TaxonG,TaxonH,TaxonI,TaxonJ,TaxonK,TaxonL,TaxonN)); 14 majorhypothesis minorhypothesis majorhypothesis 1 minorhypothesis
98 % majorhypothesis 6 TaxonG TaxonN OTU minorhypothesis pgsplicetree majorhypothesis 6 ( majorhypothesis 6.nwk ) pgsplicetree 6 input_file majorhypothesis_6.nwk MCMC pgsumtree --mode=alli --treefile=majorhypothesis_6.nwk *_bootstrap.nwk output_file [ majorincompatible_1_of_tree_1] (( TaxonA,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonH, TaxonI,TaxonJ,TaxonK,TaxonL,TaxonM,TaxonN)8.0,( TaxonO,TaxonP,TaxonG)); 2 [ minorincompatible_1_of_tree_1] (( TaxonA,TaxonB,TaxonC,TaxonD,TaxonE,TaxonF,TaxonG, TaxonH,TaxonI,TaxonJ,TaxonK,TaxonL,TaxonM)7.0,( TaxonO,TaxonP,TaxonN)); majorincompatible N of tree K --treefile K N N 2 N=1 minorincompatible majorincompatible 1 majorincompatible minorincompatible majorincompatible 1 2 minorincompatible MCMC 2 2
99 91 7 p Treefinder MrBayes5D KH SH AU Bayes factor 7.1 Treefinder Treefinder (topological constraint) TaxonA TaxonE 5 OTU TaxonA TaxonB (monophyly) (( TaxonA,TaxonB),TaxonC,TaxonD,TaxonE); (( TaxonA,TaxonB),(TaxonC,TaxonD,TaxonE)); TaxonA TaxonB TaxonA TaxonB TaxonC ((( TaxonA,TaxonB),TaxonC),TaxonD,TaxonE);
100 92 7 (positive constraint) (negative constraint) Treefinder 2 (= ) partition criterion xxx singlesearch.tl report:=reconstructphylogeny[ 2 " partition_xxx.tf", 3 SubstitutionModel ->Load[ 4 " partition_criterion_xxx.model" 5 ], 6 PartitionRates ->Load[ 7 " partition_criterion_xxx.rates" 8 ], 9 Tree ->" constraint.nwk", 10 ResolveMultifurcations ->True, 11 WithEdgeSupport ->False, 12 SearchDepth ->2, 13 AcceptFlatness ->True 14 ], 15 Oprec[ 16 20, 17 SaveReport[ 18 AsReport[ 19 report 20 ], 21 " partition_criterion_xxx_treesearch_constraint.log" 22 ], 23 Save[ 24 report 1 SubstitutionModel, 25 " partition_criterion_xxx_optimum_constraint.model" 26 ], 27 Save[ 28 report 1 PartitionRates, 29 " partition_criterion_xxx_optimum_constraint.rates" 30 ], 31 SaveTree[ 32 AsTree[ 33 report 1 Phylogeny 34 ], 35 " partition_criterion_xxx_optimum_constraint.nwk", 36 Format ->"NEWICK" 37 ] 38 ] Tree->"constraint.nwk",
101 7.2 Treefinder 93 ResolveMultifurcations->True, 2 Treefinder Kernel Load TL Script... likelihood ratchet pgtfratchet If you want to give topological constraint, specify an input file name. Otherwise, just press enter. pgtfratchet TNT 7.3 TNT pgtfratchet OTU TaxonA TaxonA OTU (TaxonD,TaxonE,(( TaxonA,TaxonB),TaxonC)); TaxonD 7.2 TaxonA TaxonB TaxonC TaxonD TaxonE OTU 7.2 Treefinder
102 KH SH AU Kishino-Hasegawa (KH test) (Kishino and Hasegawa, 1989) 3 1 ( ) Shimodaira-Hasegawa (SH test) (Shimodaira and Hasegawa, 1999) 2 ( ) (approximately unbiased [AU] test) (Shimodaira, 2002) Treefinder.log.log.log (Treefinder Report File ) Treefinder Utilities Join Report Files OK Report File Phylogears2 pgtfjoinlog 3 pgtfjoinlog input_file1 input_file2 output_file Report File Analysis Test Hypotheses... Sequence File Report File ( partition xxx.tf) Hypothesis File Report File # Replicates ( ) (AU 10 ) Criterion Likelihood OK File Export Tree... Report File Save PostScript Report File KH SH p 1 p
103 7.2 Treefinder (parametric bootstrap test) ( ) KH SH AU Treefinder Report File Report File Analysis Parametric Bootstrap Test... H0 = Report File H1 Report File # Replicates ( ) Criterion Likelihood OK Treefinder p ( ) 2 KH SH AU KH SH AU 2 KH SH AU ( )
104 MrBayes5D Treefinder TaxonA TaxonE 5 OTU TaxonA TaxonB (monophyly) NEXUS NEXUS MrBayes ( ) MrBayes > Constraint monophyly1 100=TaxonA TaxonB MrBayes > PrSet TopologyPr=Constraints(monophyly1) TaxonA TaxonB TaxonC MrBayes > Constraint monophyly1 100=TaxonA TaxonB MrBayes > Constraint monophyly2 100=TaxonA TaxonB TaxonC MrBayes > PrSet TopologyPr=Constraints(monophyly1,monophyly2) MrBayes5D Treefinder 7.4 Bayes factor Bayes factor (Kass and Raftery, 1995) MCMC Bayes factor Bayes factor Bayes factor MCMC Tracer Bayes factor (Newton and Raftery, 1994) Bayes factor 1 NEXUS constraint1.nex 2 NEXUS constraint2.nex MCMC constraint1.nex.run1.p constraint1.nex.run2.p constraint2.nex.run1.p constraint2.nex.run2.p 4 burn-in (
105 7.4 Bayes factor 97 ) Phylogears2 pgmbburninparam 2 burn-in burn-in constraint1 param.txt constraint2 param.txt pgmbburninparam --burnin=10001 constraint1.nex.run1.p constraint1_param.txt pgmbburninparam --burnin= append constraint1.nex.run2.p constraint1_param.txt pgmbburninparam --burnin=15001 constraint2.nex.run1.p constraint2_param.txt pgmbburninparam --burnin= append constraint2.nex.run2.p constraint2_param.txt burn-in Tracer File Import Trace File... constraint1 param.txt constraint2 param.txt Trace Files Burn-In 0 Analysis Calculate Bayes Factors... Likelihood trace LnL Calculate harmonic mean only (no smoothing) Bootstrap replicates 1000 Show ln Bayes Factors Trace ln Bayes factor ln Bayes factor 7.1 (Kass and Raftery, 1995) 7.1 Bayes factor ln Bayes factor MrBayes5D 2 MCMC 2 MCMC Bayes factor 2 MCMC Bayes factor Bayes factor
106
107 , Sudhir Kumar ISBN Kumar Ziheng Yang ISBN Yang
108 100 8 Inferring Phylogenies Joseph Felsenstein Sinauer Associates Inc. ISBN Felsenstein The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing Philippe Lemey, Marco Salemi, Anne-Mieke Vandamme Cambridge University Press ISBN ,,, ISBN AIC KH SH AU
109 8.3 UNIX 101 ISBN MCMC MrBayes II,,,,, ISBN MCMC 8.3 UNIX UNIX Windows UNIX Cygwin Linux Ubuntu Linux MacOS X UNIX CD DVD Web UNIX Gentoo Linux UNIX UNIX SSH GNU screen tmux Web
110 102 8 Windows UNIX Cygwin ISBN Windows UNIX Cygwin UNIX ISBN Ubuntu Linux ISBN Ubuntu ISBN MacOS X UNIX UNIX ISBN Unix for Mac OS X Dave Taylor
111 8.3 UNIX 103 ISBN IDG ISBN UNIX bash UNIX, ISBN
112
113 105 Ababneh, F., Jermiin, L. S., Ma, C., and Robinson, J., 2006, Matched-pairs tests of homogeneity with applications to homologous nucleotide sequences, Bioinformatics, 22, Abascal, F., Posada, D., and Zardoya, R., 2007, MtArt: a new model of amino acid replacement for Arthropoda, Molecular Biology and Evolution, 24, 1 5. Adachi, J. and Hasegawa, M., 1996, MOLPHY version 2.3: programs for molecular phylogenetics based in maximum likelihood, Computer Science Monographs, 28, Adachi, J., Waddell, P. J., Martin, W., and Hasegawa, M., 2000, Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA, Journal of Molecular Evolution, 50, Akaike, H., 1974, New look at statistical-model identification, IEEE Transactions on Automatic Control, 19, Altekar, G., Dwarkadas, S., Huelsenbeck, J. P., and Ronquist, F., 2004, Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference, Bioinformatics, 20, Blanquart, S. and Lartillot, N., 2006, A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution, Molecular Biology and Evolution, 23, No. 11, , Nov., 2008, A site- and time-heterogeneous model of amino acid replacement, Molecular Biology and Evolution, 25, No. 5, , May. Boussau, B. and Gouy, M., 2006, Efficient likelihood computations with nonreversible models of evolution, Systematic Biology, 55, No. 5, , Oct. Cao, Y., Janke, A., Waddell, P. J., Westerman, M., Takenaka, O., Murata, S., Okada, N., Pääbo, S., and Hasegawa, M., 1998, Conflict among individual mitochondrial proteins in resolving the phylogeny of eutherian orders., Journal of Molecular Evolution, 47, Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T., 2009, trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses, Bioinformatics, 25, No. 15, , Aug. Castresana, J., 2000, Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis, Molecular Biology and Evolution, 17, No. 4, , Apr. Cochran, W. G., 1954, Some methods for strengthening the common χ 2 tests, Biometrics, 10, Criscuolo, A. and Gribaldo, S., 2010, BMGE (Block Mapping and Gathering with Entropy): a new software for
114 106 selection of phylogenetic informative regions from multiple sequence alignments, BMC Evolutionary Biology, 10, 210. Dayhoff, M. O., Schwartz, R. M., and Orcutt, B. C., 1978, A model of evolutionary change in proteins, Vol. 5, Suppl. 3, in Dayhoff, M. O. ed. Atlas of Protein Sequence Structure: National Biomedical Research Foundation, Dimmic, M. W., Rest, J. S., Mindell, D. P., and Goldstein, R. A., 2002, rtrev: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny, Journal of Molecular Evolution, 55, Edgar, R. C., 2004, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Research, 32, No. 5, Felsenstein, J., 1981, Evolutionary trees from DNA sequencies - a maximum-likelihood approach, Journal of Molecular Evolution, 17, , 1985, Confidence-limits on phylogenies - an approach using the bootstrap, Evolution, 39, Fleissner, R., Metzler, D., and von Haeseler, A., 2005, Simultaneous statistical multiple alignment and phylogeny reconstruction, Systematic Biology, 54, Hastings, W. K., 1970, Monte Carlo sampling methods using Markov chains and their applications, Biometrika, 57, Henikoff, S. and Henikoff, J. G., 1992, Amino acid substitution matrices from protein blocks, Proceedings of the National Academy of Sciences of the United States of America, 89, Hrdy, I., Hirt, R. P., Dolezal, P., Bardonová, L., Foster, P. G., Tachezy, J., and Embley, T. M., 2004, Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I, Nature, 432, No. 7017, , Dec. Jobb, G., 2008, Treefinder version of April 2008, Software distributed by the author at Jobb, G., von Haeseler, A., and Strimmer, K., 2004, Treefinder: a powerful graphical analysis environment for molecular phylogenetics, BMC Evolutionary Biology, 4, 18. Jones, D. T., Taylor, W. R., and Thornton, J. M., 1992, The rapid generation of mutation data matrices from protein sequences, Computer Applications in the Biosciences, 8, Jukes, T. H. and Cantor, C. R., 1969, Evolution of protein molecules, in Munro, H. N. ed. Mammalian protein metabolism, New York: Academic Press, Kass, R. E. and Raftery, A. E., 1995, Bayes Factors, Journal of the American Statistical Association, 90, Kass, R. E., Carlin, B. P., Gelman, A., and Neal, R., 1998, Markov chain Monte Carlo in practice: a roundtable discussion, American Statistician, 52, Katoh, K., Kuma, K., Toh, H., and Miyata, T., 2005, MAFFT version 5: improvement in accuracy of multiple sequence alignment, Nucleic Acids Research, 33, Kimura, M., 1980, A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences, Journal of Molecular Evolution, 16,
115 107, 1983, The neutral theory of molecular evolution: Cambridge University Press. Kishino, H. and Hasegawa, M., 1989, Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea, Journal of Molecular Evolution, 29, Kosiol, C. and Goldman, N., 2005, Different versions of the Dayhoff rate matrix, Molecular Biology and Evolution, 22, Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G., 2007, Clustal W and Clustal X version 2.0, Bioinformatics, 23, No. 21, , Nov. Le, S. Q. and Gascuel, O., 2008, An improved general amino acid replacement matrix, Molecular Biology and Evolution, 25, Lunter, G., Miklós, I., Drummond, A., Jensen, J. L., and Hein, J., 2005, Bayesian coestimation of phylogeny and sequence alignment, BMC Bioinformatics, 6, 83. Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., and Teller, A. H., 1953, Equation of state calculations by fast computing machines, Journal of Chemical Physics, 21, Müller, T. and Vingron, M., 2000, Modeling amino acid replacement, Journal of Computational Biology, 7, Newton, M. A. and Raftery, A. E., 1994, Approximate Bayesian inference with the weighted likelihood bootstrap, Journal of the Royal Statistical Society, 56, Nickle, D. C., Heath, L., Jensen, M. A., Gilbert, P. B., Mullins, J. I., and Pond, S. L. K., 2007, HIV-specific probabilistic models of protein evolution, PLoS ONE, 2, e503. Nixon, K. C., 1999, The parsimony ratchet : a new method for rapid parsimony analysis, Cladistics, 15, Posada, D. and Crandall, K. A., 1998, Modeltest: testing the model of DNA substitution, Bioinformatics, 14, Redelings, B. D. and Suchard, M. A., 2005, Joint Bayesian estimation of alignment and phylogeny, Systematic Biology, 54, Ronquist, F. and Huelsenbeck, J. P., 2003, MrBayes 3: Bayesian phylogenetic inference under mixed models, Bioinformatics, 19, Ronquist, F., Huelsenbeck, J. P., and van der Mark, P., 2005, MrBayes 3.1 Manual 5/26/2005, Distributed at Rota-Stabelli, O., Yang, Z., and Telford, M. J., 2009, MtZoa: a general mitochondrial amino acid substitutions model for animal evolutionary studies, Molecular Phylogenetics and Evolution, 52, No. 1, , Jul. Saitou, N. and Nei, M., 1987, The neighbor-joining method: a new method for reconstructing phylogenetics trees, Molecular Biology and Evolution, 4, Schwarz, G., 1978, Estimating the dimension of a model, Annals of Statistics, 6,
116 108 Shimodaira, H., 2002, An approximately unbiased test of phylogenetic tree selection, Systematic Biology, 51, Shimodaira, H. and Hasegawa, M., 1999, Multiple comparisons of log-likelihoods with applications to phylogenetic inference, Molecular Biology and Evolution, 16, Stamatakis, A., 2006, RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models, Bioinformatics, 22, Sugiura, N., 1978, Further analysis of the data by Akaike s information criterion and the finite corrections, Communications in Statistics: Theory and Methods, A7, Swofford, D. L., 2003, PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4, Sunderland, Massachusetts: Sinauer Associates. Swofford, D. L. and Begle, D. P., 1993, PAUP: Phylogenetic Analysis Using Parsimony, Ver.3.1. User s Manual: Laboratory of Molecular Systematics, Smithonian Institution. Talavera, G. and Castresana, J., 2007, Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments, Systematic Biology, 56, No. 4, , Aug. Tanabe, A. S., 2007, Kakusan: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data, Molecular Ecology Notes, 7, Tavaré, S., 1986, Some probabilistic and statistical problems in the analysis of DNA sequences, Lectures on Mathematics in the Life Sciences, 17, Veerassamy, S., Smith, A., and Tillier, E. R. M., 2003, A transition probability model for amino acid substitutions from blocks., Journal of Computational Biology, 10, Vos, R. A., 2003, Accelerated likelihood surface exploration: the likelihood ratchet, Systematic Biology, 52, Whelan, S. and Goldman, N., 2001, A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach, Molecular Biology and Evolution, 18, Woese, C. R., Achenbach, L., Rouviere, P., and Mandelco, L., 1991, Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain composition-induced artifacts, Systematic and Applied Microbiology, 14, No. 4, Yang, Z., 1993, Maximum likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites, Molecular Biology and Evolution, 10, , 1994, Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods, Journal of Molecular Evolution, 39, , 1995, A space-time process model for the evolution of DNA sequences, Genetics, 139,
分子系統学演習
 2013 3 26 i 1 0 3 0.1 Windows............................................ 3 0.2 Mac OS X........................................... 5 0.3 Linux.............................................. 7 1 9 1.1................................
2013 3 26 i 1 0 3 0.1 Windows............................................ 3 0.2 Mac OS X........................................... 5 0.3 Linux.............................................. 7 1 9 1.1................................
分子系統学演習
 2015/10/20 i 1 3 0 5 0.1 Windows............................................... 5 0.2 Mac OS X............................................... 7 0.3 Linux.................................................
2015/10/20 i 1 3 0 5 0.1 Windows............................................... 5 0.2 Mac OS X............................................... 7 0.3 Linux.................................................
untitled
 VQT3B82-1 DMP-BDT110 μ μ μ 2 VQT3B82 ÇÕÇ¹Ç Ç +- VQT3B82 3 4 VQT3B82 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 ij SD 1 2 3 4 5 6 7 8 Í VQT3B82 5 BD DVD CD SD USB 6 VQT3B82 2 ALL 1 2 4 VQT3B82 7
VQT3B82-1 DMP-BDT110 μ μ μ 2 VQT3B82 ÇÕÇ¹Ç Ç +- VQT3B82 3 4 VQT3B82 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 ij SD 1 2 3 4 5 6 7 8 Í VQT3B82 5 BD DVD CD SD USB 6 VQT3B82 2 ALL 1 2 4 VQT3B82 7
 class Cpd MW = { 'C'=>12.011, 'H'=>1.00794, 'N'=>14.00674, 'O' => 15.9994, 'P' => 30.973762 } def initialize @comp = Hash.new attr_accessor :name, :definition, :formula # formula def composition @formula.scan(/([a-z]+)(\d+)/)
class Cpd MW = { 'C'=>12.011, 'H'=>1.00794, 'N'=>14.00674, 'O' => 15.9994, 'P' => 30.973762 } def initialize @comp = Hash.new attr_accessor :name, :definition, :formula # formula def composition @formula.scan(/([a-z]+)(\d+)/)
 -5 DMP-BV300 μ μ l μ l l +- l l j j j l l l l l l l l l l l l l Ë l l l l l l l l l l l l l l l l l l l l l l l BD DVD CD SD USB 2 ALL 1 2 4 l l DETACH ATTACH RELEASE DETACH ATTACH DETACH ATTACH RELEASE
-5 DMP-BV300 μ μ l μ l l +- l l j j j l l l l l l l l l l l l l Ë l l l l l l l l l l l l l l l l l l l l l l l BD DVD CD SD USB 2 ALL 1 2 4 l l DETACH ATTACH RELEASE DETACH ATTACH DETACH ATTACH RELEASE
Sequencher 4.9 Confidence score Clustal Clustal ClustalW Sequencher ClustalW Windows Macintosh motif confidence Sequencher V4.9 Trim Ends Without Prev
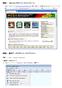
 2009 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074 (fax) www.genecodes.com [email protected]
2009 Gene Codes Corporation Gene Codes Corporation 775 Technology Drive, Ann Arbor, MI 48108 USA 1.800.497.4939 (USA) +1.734.769.7249 (elsewhere) +1.734.769.7074 (fax) www.genecodes.com [email protected]
Introduction Purpose This training course demonstrates the use of the High-performance Embedded Workshop (HEW), a key tool for developing software for
 Introduction Purpose This training course demonstrates the use of the High-performance Embedded Workshop (HEW), a key tool for developing software for embedded systems that use microcontrollers (MCUs)
Introduction Purpose This training course demonstrates the use of the High-performance Embedded Workshop (HEW), a key tool for developing software for embedded systems that use microcontrollers (MCUs)
 μ μ DMR-BZT700 DMR-BZT600 μ TM VQT3C03-2B ! ! l l l [HDD] [BD-RE] [BD-R] [DVD-V] [BD-V] [RAM] [CD] [SD] [-R] [USB] [-RW] [RAM AVCREC ] [-R AVCREC ] [RAM VR ][-R VR ] [-RW VR ] [-R V ] [-RW V ] [DVD-V]
μ μ DMR-BZT700 DMR-BZT600 μ TM VQT3C03-2B ! ! l l l [HDD] [BD-RE] [BD-R] [DVD-V] [BD-V] [RAM] [CD] [SD] [-R] [USB] [-RW] [RAM AVCREC ] [-R AVCREC ] [RAM VR ][-R VR ] [-RW VR ] [-R V ] [-RW V ] [DVD-V]
VQT3B86-4 DMP-HV200 DMP-HV150 μ μ l μ
 -4 DMP-HV200 DMP-HV150 μ μ l μ [DMP-HV200] l [DMP-HV200] l +- l l j j j[dmp-hv200] l l l [DMP-HV200] l l l l [DMP-HV200] l [DMP-HV200] l l [DMP-HV200] l [DMP-HV200] [DMP-HV150] l l Ë l l l l l l l l l
-4 DMP-HV200 DMP-HV150 μ μ l μ [DMP-HV200] l [DMP-HV200] l +- l l j j j[dmp-hv200] l l l [DMP-HV200] l l l l [DMP-HV200] l [DMP-HV200] l l [DMP-HV200] l [DMP-HV200] [DMP-HV150] l l Ë l l l l l l l l l
 VQT3A26-1 DMR-T2000R μ μ μ ! R ! l l l [HDD] [BD-RE] [BD-R] [BD-V] [RAM] [-R] [-R]DL] [-RW] [DVD-V] [CD] [SD] [USB] [RAM AVCREC ] [-R AVCREC ] [-R]DL AVCREC ] [RAM VR ][-R VR ] [-R]DL VR ] [-RW VR ]
VQT3A26-1 DMR-T2000R μ μ μ ! R ! l l l [HDD] [BD-RE] [BD-R] [BD-V] [RAM] [-R] [-R]DL] [-RW] [DVD-V] [CD] [SD] [USB] [RAM AVCREC ] [-R AVCREC ] [-R]DL AVCREC ] [RAM VR ][-R VR ] [-R]DL VR ] [-RW VR ]
untitled
 TZ-BDT910M TZ-BDT910F TZ-BDT910P μ μ μ μ TM VQT3F51-1 l l l [HDD] [BD-RE] [BD-R] [DVD-V] [BD-V] [RAM] [CD] [SD] [-R] [USB] [-RW] [RAM AVCREC ] [-R AVCREC ] [RAM VR ][-R VR ] [-RW VR ] [-R V ] [-RW
TZ-BDT910M TZ-BDT910F TZ-BDT910P μ μ μ μ TM VQT3F51-1 l l l [HDD] [BD-RE] [BD-R] [DVD-V] [BD-V] [RAM] [CD] [SD] [-R] [USB] [-RW] [RAM AVCREC ] [-R AVCREC ] [RAM VR ][-R VR ] [-RW VR ] [-R V ] [-RW
Kaplan-Meierプロットに付加情報を追加するマクロの作成
 Kaplan-Meier 1, 2,3 1 2 3 A SAS macro for extended Kaplan-Meier plots Kengo Nagashima 1, Yasunori Sato 2,3 1 Department of Parmaceutical Technochemistry, Josai University 2 School of Medicine, Chiba University
Kaplan-Meier 1, 2,3 1 2 3 A SAS macro for extended Kaplan-Meier plots Kengo Nagashima 1, Yasunori Sato 2,3 1 Department of Parmaceutical Technochemistry, Josai University 2 School of Medicine, Chiba University
分子系統樹推定の落とし穴と回避法 筑波大 生命環境 田辺晶史
 分子系統樹推定の落とし穴と回避法 筑波大 生命環境 田辺晶史 http://www.fifthdimension.jp/wiki.cgi http://www.fifthdimension.jp/documents/molphytextbook/ 分子系統樹推定 の 落とし穴 とは データが 仮定 を満たしていない 仮定その1 相同 である 相同 非相同 相同 相同 同一の祖先形質 に由来する
分子系統樹推定の落とし穴と回避法 筑波大 生命環境 田辺晶史 http://www.fifthdimension.jp/wiki.cgi http://www.fifthdimension.jp/documents/molphytextbook/ 分子系統樹推定 の 落とし穴 とは データが 仮定 を満たしていない 仮定その1 相同 である 相同 非相同 相同 相同 同一の祖先形質 に由来する
Microsoft Word - Meta70_Preferences.doc
 Image Windows Preferences Edit, Preferences MetaMorph, MetaVue Image Windows Preferences Edit, Preferences Image Windows Preferences 1. Windows Image Placement: Acquire Overlay at Top Left Corner: 1 Acquire
Image Windows Preferences Edit, Preferences MetaMorph, MetaVue Image Windows Preferences Edit, Preferences Image Windows Preferences 1. Windows Image Placement: Acquire Overlay at Top Left Corner: 1 Acquire
Introduction Purpose This training course describes the configuration and session features of the High-performance Embedded Workshop (HEW), a key tool
 Introduction Purpose This training course describes the configuration and session features of the High-performance Embedded Workshop (HEW), a key tool for developing software for embedded systems that
Introduction Purpose This training course describes the configuration and session features of the High-performance Embedded Workshop (HEW), a key tool for developing software for embedded systems that
untitled
 FutureNet Microsoft Corporation Microsoft Windows Windows 95 Windows 98 Windows NT4.0 Windows 2000, Windows XP, Microsoft Internet Exproler (1) (2) (3) COM. (4) (5) ii ... 1 1.1... 1 1.2... 3 1.3... 6...
FutureNet Microsoft Corporation Microsoft Windows Windows 95 Windows 98 Windows NT4.0 Windows 2000, Windows XP, Microsoft Internet Exproler (1) (2) (3) COM. (4) (5) ii ... 1 1.1... 1 1.2... 3 1.3... 6...
バクテリアゲノム解析
 GCCGTAGCTACCTTTACAATA GCCGTAGCT AGCTACC GCTACCTTT CCTTTAC CTTTACAATA GCCG CCGT CGTA GTAG TAGC AGCT AGCT GCTA CTAC TACC GCTA CTAC TACC ACCT CCTT CTTT CCTT CTTT TTTA TTAC CTTT TTTA TTAC TACA ACAA CAAT AATA
GCCGTAGCTACCTTTACAATA GCCGTAGCT AGCTACC GCTACCTTT CCTTTAC CTTTACAATA GCCG CCGT CGTA GTAG TAGC AGCT AGCT GCTA CTAC TACC GCTA CTAC TACC ACCT CCTT CTTT CCTT CTTT TTTA TTAC CTTT TTTA TTAC TACA ACAA CAAT AATA
 HARK Designer Documentation 0.5.0 HARK support team 2013 08 13 Contents 1 3 2 5 2.1.......................................... 5 2.2.............................................. 5 2.3 1: HARK Designer.................................
HARK Designer Documentation 0.5.0 HARK support team 2013 08 13 Contents 1 3 2 5 2.1.......................................... 5 2.2.............................................. 5 2.3 1: HARK Designer.................................
分子系統樹作成方法
 実 習 1: MEGA6 のダウンロードとインストール MEGA の Web サイトは http://www.megasoftware.net/( 下 図 ) 正 式 には 左 側 の[Windows]ボタンをクリックし 名 前 とメールアドレスを 入 力 して[Submit Request]をクリックすると ダウンロード 用 のアドレスがメールで 送 られる 実 習 2: 配 列 データのダウンロードとアライメント
実 習 1: MEGA6 のダウンロードとインストール MEGA の Web サイトは http://www.megasoftware.net/( 下 図 ) 正 式 には 左 側 の[Windows]ボタンをクリックし 名 前 とメールアドレスを 入 力 して[Submit Request]をクリックすると ダウンロード 用 のアドレスがメールで 送 られる 実 習 2: 配 列 データのダウンロードとアライメント
外部SQLソース入門
 Introduction to External SQL Sources 外部 SQL ソース入門 3 ESS 3 ESS : 4 ESS : 4 5 ESS 5 Step 1:... 6 Step 2: DSN... 6 Step 3: FileMaker Pro... 6 Step 4: FileMaker Pro 1. 6 Step 5:... 6 Step 6: FileMaker Pro...
Introduction to External SQL Sources 外部 SQL ソース入門 3 ESS 3 ESS : 4 ESS : 4 5 ESS 5 Step 1:... 6 Step 2: DSN... 6 Step 3: FileMaker Pro... 6 Step 4: FileMaker Pro 1. 6 Step 5:... 6 Step 6: FileMaker Pro...
Introduction Purpose This course explains how to use Mapview, a utility program for the Highperformance Embedded Workshop (HEW) development environmen
 Introduction Purpose This course explains how to use Mapview, a utility program for the Highperformance Embedded Workshop (HEW) development environment for microcontrollers (MCUs) from Renesas Technology
Introduction Purpose This course explains how to use Mapview, a utility program for the Highperformance Embedded Workshop (HEW) development environment for microcontrollers (MCUs) from Renesas Technology
EPSON ES-D200 パソコンでのスキャンガイド
 NPD4271-00 ...4...7 EPSON Scan... 7...11 PDF...12 / EPSON Scan...13 EPSON Scan...13 EPSON Scan...14 EPSON Scan...14 EPSON Scan...15 Epson Event Manager...16 Epson Event Manager...16 Epson Event Manager...16
NPD4271-00 ...4...7 EPSON Scan... 7...11 PDF...12 / EPSON Scan...13 EPSON Scan...13 EPSON Scan...14 EPSON Scan...14 EPSON Scan...15 Epson Event Manager...16 Epson Event Manager...16 Epson Event Manager...16
DS-30
 NPD4633-00 JA ...6... 6... 6... 6... 6... 7... 7... 7... 7... 8... 8...9...10...11...11...13 Document Capture Pro Windows...13 EPSON Scan Mac OS X...14 SharePoint Windows...16 Windows...16...17 Document
NPD4633-00 JA ...6... 6... 6... 6... 6... 7... 7... 7... 7... 8... 8...9...10...11...11...13 Document Capture Pro Windows...13 EPSON Scan Mac OS X...14 SharePoint Windows...16 Windows...16...17 Document
EPSON Easy Interactive Tools Ver.4.2 Operation Guide
 Easy Interactive Tools Ver.4.2 Easy Interactive Tools Ver.4.2 Easy Interactive Tools Easy Interactive Tools Easy Interactive Pen s p.12 s p.12 s p.11 s p.18 s p.20 s p.29 Easy Interactive Tools PowerPoint
Easy Interactive Tools Ver.4.2 Easy Interactive Tools Ver.4.2 Easy Interactive Tools Easy Interactive Tools Easy Interactive Pen s p.12 s p.12 s p.11 s p.18 s p.20 s p.29 Easy Interactive Tools PowerPoint
はじめに
 IT 1 NPO (IPEC) 55.7 29.5 Web TOEIC Nice to meet you. How are you doing? 1 type (2002 5 )66 15 1 IT Java (IZUMA, Tsuyuki) James Robinson James James James Oh, YOU are Tsuyuki! Finally, huh? What's going
IT 1 NPO (IPEC) 55.7 29.5 Web TOEIC Nice to meet you. How are you doing? 1 type (2002 5 )66 15 1 IT Java (IZUMA, Tsuyuki) James Robinson James James James Oh, YOU are Tsuyuki! Finally, huh? What's going
エレクトーンのお客様向けiPhone/iPad接続マニュアル
 / JA 1 2 3 4 USB TO DEVICE USB TO DEVICE USB TO DEVICE 5 USB TO HOST USB TO HOST USB TO HOST i-ux1 6 7 i-ux1 USB TO HOST i-mx1 OUT IN IN OUT OUT IN OUT IN i-mx1 OUT IN IN OUT OUT IN OUT IN USB TO DEVICE
/ JA 1 2 3 4 USB TO DEVICE USB TO DEVICE USB TO DEVICE 5 USB TO HOST USB TO HOST USB TO HOST i-ux1 6 7 i-ux1 USB TO HOST i-mx1 OUT IN IN OUT OUT IN OUT IN i-mx1 OUT IN IN OUT OUT IN OUT IN USB TO DEVICE
PX-504A
 NPD4537-00 ...6... 6... 9 Mac OS X...10 Mac OS X v10.5.x v10.6.x...10 Mac OS X v10.4.11...13...15...16...16...18...19...20!ex...20 /...21 P.I.F. PRINT Image Framer...21...22...26...26...27...27...27...31
NPD4537-00 ...6... 6... 9 Mac OS X...10 Mac OS X v10.5.x v10.6.x...10 Mac OS X v10.4.11...13...15...16...16...18...19...20!ex...20 /...21 P.I.F. PRINT Image Framer...21...22...26...26...27...27...27...31
EPSON PX-503A ユーザーズガイド
 NPD4296-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...21...22...23!ex...23 /...24 P.I.F. PRINT Image Framer...24...25...28...28...29...29...30...33
NPD4296-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...21...22...23!ex...23 /...24 P.I.F. PRINT Image Framer...24...25...28...28...29...29...30...33
PX-403A
 NPD4403-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...21...22!ex...22 /...23 P.I.F. PRINT Image Framer...23...24...27...27...28...28...28...32 Web...32...32...35...35...35...37...37...37...39...39...40...43...46
NPD4403-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...21...22!ex...22 /...23 P.I.F. PRINT Image Framer...23...24...27...27...28...28...28...32 Web...32...32...35...35...35...37...37...37...39...39...40...43...46
インターネット接続ガイド v110
 1 2 1 2 3 3 4 5 6 4 7 8 5 1 2 3 6 4 5 6 7 7 8 8 9 9 10 11 12 10 13 14 11 1 2 12 3 4 13 5 6 7 8 14 1 2 3 4 < > 15 5 6 16 7 8 9 10 17 18 1 2 3 19 1 2 3 4 20 U.R.G., Pro Audio & Digital Musical Instrument
1 2 1 2 3 3 4 5 6 4 7 8 5 1 2 3 6 4 5 6 7 7 8 8 9 9 10 11 12 10 13 14 11 1 2 12 3 4 13 5 6 7 8 14 1 2 3 4 < > 15 5 6 16 7 8 9 10 17 18 1 2 3 19 1 2 3 4 20 U.R.G., Pro Audio & Digital Musical Instrument
fx-9860G Manager PLUS_J
 fx-9860g J fx-9860g Manager PLUS http://edu.casio.jp k 1 k III 2 3 1. 2. 4 3. 4. 5 1. 2. 3. 4. 5. 1. 6 7 k 8 k 9 k 10 k 11 k k k 12 k k k 1 2 3 4 5 6 1 2 3 4 5 6 13 k 1 2 3 1 2 3 1 2 3 1 2 3 14 k a j.+-(),m1
fx-9860g J fx-9860g Manager PLUS http://edu.casio.jp k 1 k III 2 3 1. 2. 4 3. 4. 5 1. 2. 3. 4. 5. 1. 6 7 k 8 k 9 k 10 k 11 k k k 12 k k k 1 2 3 4 5 6 1 2 3 4 5 6 13 k 1 2 3 1 2 3 1 2 3 1 2 3 14 k a j.+-(),m1
NSR-500 Create DVD Installer Procedures
 Creating NSR-500 DVD Installer Overview This document describes how to create DVD installer for the NSR-500 series. Applicable Model NSR-500 Series To be required * Windows (XP, Vista or 7) installed PC
Creating NSR-500 DVD Installer Overview This document describes how to create DVD installer for the NSR-500 series. Applicable Model NSR-500 Series To be required * Windows (XP, Vista or 7) installed PC
EPSON EP-803A/EP-803AW ユーザーズガイド
 NPD4293-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...22...23...24!ex...24 /...25 P.I.F. PRINT Image Framer...25...26...30...30...31...31...31...35
NPD4293-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...22...23...24!ex...24 /...25 P.I.F. PRINT Image Framer...25...26...30...30...31...31...31...35
25 II :30 16:00 (1),. Do not open this problem booklet until the start of the examination is announced. (2) 3.. Answer the following 3 proble
 25 II 25 2 6 13:30 16:00 (1),. Do not open this problem boolet until the start of the examination is announced. (2) 3.. Answer the following 3 problems. Use the designated answer sheet for each problem.
25 II 25 2 6 13:30 16:00 (1),. Do not open this problem boolet until the start of the examination is announced. (2) 3.. Answer the following 3 problems. Use the designated answer sheet for each problem.
HA8000シリーズ ユーザーズガイド ~BIOS編~ HA8000/RS110/TS10 2013年6月~モデル
 P1E1M01500-3 - - - LSI MegaRAID SAS-MFI BIOS Version x.xx.xx (Build xxxx xx, xxxx) Copyright (c) xxxx LSI Corporation HA -0 (Bus xx Dev
P1E1M01500-3 - - - LSI MegaRAID SAS-MFI BIOS Version x.xx.xx (Build xxxx xx, xxxx) Copyright (c) xxxx LSI Corporation HA -0 (Bus xx Dev
EPSON EP-703A ユーザーズガイド
 NPD4295-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...22...23...24!ex...24 /...25 P.I.F. PRINT Image Framer...25...26...29...30...30...31...31...34
NPD4295-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...22...23...24!ex...24 /...25 P.I.F. PRINT Image Framer...25...26...29...30...30...31...31...34
iPhone/iPad接続マニュアル
 / JA 2 3 USB 4 USB USB i-ux1 USB i-ux1 5 6 i-mx1 THRU i-mx1 THRU 7 USB THRU 1 2 3 4 1 2 3 4 5 8 1 1 9 2 1 2 10 1 2 2 6 7 11 1 2 3 4 5 6 7 8 12 1 2 3 4 5 6 13 14 15 WPA Supplicant Copyright 2003-2009, Jouni
/ JA 2 3 USB 4 USB USB i-ux1 USB i-ux1 5 6 i-mx1 THRU i-mx1 THRU 7 USB THRU 1 2 3 4 1 2 3 4 5 8 1 1 9 2 1 2 10 1 2 2 6 7 11 1 2 3 4 5 6 7 8 12 1 2 3 4 5 6 13 14 15 WPA Supplicant Copyright 2003-2009, Jouni
基本操作ガイド
 HT7-0199-000-V.5.0 1. 2. 3. 4. 5. 6. 7. 8. 9. Copyright 2004 CANON INC. ALL RIGHTS RESERVED 1 2 3 1 1 2 3 4 1 2 1 2 3 1 2 3 1 2 3 1 2 3 4 1 2 3 4 1 2 3 4 5 AB AB Step 1 Step
HT7-0199-000-V.5.0 1. 2. 3. 4. 5. 6. 7. 8. 9. Copyright 2004 CANON INC. ALL RIGHTS RESERVED 1 2 3 1 1 2 3 4 1 2 1 2 3 1 2 3 1 2 3 1 2 3 4 1 2 3 4 1 2 3 4 5 AB AB Step 1 Step
Mrbayesのダウンロード MrbayesのHP(MrBayes: Bayesian Inference of Phylogeny)アドレスは
 MrBayes の使い方 目 次 1 使用法概説 1.1 Mrbayes のダウンロードとインストール 1 1.2 データファイルの作成 2 1.3 データファイルの読み込みと事前設定 3 1.4 解析の実行 5 2 塩基の置換モデルと置換率の決定 MrAIC の使用法 2.1 準備 6 2.2 データファイルの用意 6 2.3 MrAIC の実行 7 2.4 実行結果 7 3 GAP コーディング
MrBayes の使い方 目 次 1 使用法概説 1.1 Mrbayes のダウンロードとインストール 1 1.2 データファイルの作成 2 1.3 データファイルの読み込みと事前設定 3 1.4 解析の実行 5 2 塩基の置換モデルと置換率の決定 MrAIC の使用法 2.1 準備 6 2.2 データファイルの用意 6 2.3 MrAIC の実行 7 2.4 実行結果 7 3 GAP コーディング
操作ガイド(本体操作編)
 J QT5-0571-V03 1 ...5...10...11...11...11...12...12...15...21...21...22...25...27...28...33...37...40...47...48...54...60...64...64...68...69...70...70...71...72...73...74...75...76...77 2 ...79...79...80...81...82...83...95...98
J QT5-0571-V03 1 ...5...10...11...11...11...12...12...15...21...21...22...25...27...28...33...37...40...47...48...54...60...64...64...68...69...70...70...71...72...73...74...75...76...77 2 ...79...79...80...81...82...83...95...98
EP-704A
 NPD4533-01 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.11...15...18...19...19...22...23...24!ex...24 /...25 P.I.F. PRINT Image Framer...25...26...29...29...30...30...31...34
NPD4533-01 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.11...15...18...19...19...22...23...24!ex...24 /...25 P.I.F. PRINT Image Framer...25...26...29...29...30...30...31...34
PX-434A/PX-404A
 NPD4534-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.11...15...18...19...19...21...22!ex...22 /...23 P.I.F. PRINT Image Framer...23...24...26...27...27...28...28...31 Web...31...31...35...35...35...37...37...37...39...39...40...43...48
NPD4534-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.11...15...18...19...19...21...22!ex...22 /...23 P.I.F. PRINT Image Framer...23...24...26...27...27...28...28...31 Web...31...31...35...35...35...37...37...37...39...39...40...43...48
DS-70000/DS-60000/DS-50000
 NPD4647-02 JA ...5...7...8 ADF...9... 9 ADF...10...11...13...15 Document Capture Pro Windows...15 EPSON Scan Mac OS X...16 SharePoint Windows...18 Windows...18...19 Windows...19 Mac OS X...19...20...23...23
NPD4647-02 JA ...5...7...8 ADF...9... 9 ADF...10...11...13...15 Document Capture Pro Windows...15 EPSON Scan Mac OS X...16 SharePoint Windows...18 Windows...18...19 Windows...19 Mac OS X...19...20...23...23
2
 NSCP-W61 08545-00U60 2 3 4 5 6 7 8 9 10 11 12 1 2 13 7 3 4 8 9 5 6 10 7 14 11 15 12 13 16 17 14 15 1 5 2 3 6 4 16 17 18 19 2 1 20 1 21 2 1 2 1 22 23 1 2 3 24 1 2 1 2 3 3 25 1 2 3 4 1 2 26 3 4 27 1 1 28
NSCP-W61 08545-00U60 2 3 4 5 6 7 8 9 10 11 12 1 2 13 7 3 4 8 9 5 6 10 7 14 11 15 12 13 16 17 14 15 1 5 2 3 6 4 16 17 18 19 2 1 20 1 21 2 1 2 1 22 23 1 2 3 24 1 2 1 2 3 3 25 1 2 3 4 1 2 26 3 4 27 1 1 28
ScanFront300/300P セットアップガイド
 libtiff Copyright (c) 1988-1996 Sam Leffler Copyright (c) 1991-1996 Silicon Graphics, Inc. Permission to use, copy, modify, distribute, and sell this software and its documentation for any purpose is hereby
libtiff Copyright (c) 1988-1996 Sam Leffler Copyright (c) 1991-1996 Silicon Graphics, Inc. Permission to use, copy, modify, distribute, and sell this software and its documentation for any purpose is hereby
PX-673F
 NPD4385-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...21...22...23!ex...23 /...24 P.I.F. PRINT Image Framer...24...25...28...29...29...30...30...33
NPD4385-00 ...6... 6...10 Mac OS X...11 Mac OS X v10.5.x v10.6.x...11 Mac OS X v10.4.x...15...18...19...19...21...22...23!ex...23 /...24 P.I.F. PRINT Image Framer...24...25...28...29...29...30...30...33
MINI2440マニュアル
 Open-JTAG LPC2388 + GCC + Eclipse http://www.csun.co.jp [email protected] Ver1.4 2009/7/31 LPC2388 OpenJTAG copyright@2009 http://www.csun.co.jp [email protected] 1 ...3 ARM...4...5...6 4.2 OpenJTAG...6 4.2...8
Open-JTAG LPC2388 + GCC + Eclipse http://www.csun.co.jp [email protected] Ver1.4 2009/7/31 LPC2388 OpenJTAG copyright@2009 http://www.csun.co.jp [email protected] 1 ...3 ARM...4...5...6 4.2 OpenJTAG...6 4.2...8
RT-PCR プロトコール.PDF
 Real -Time RT-PCR icycler iq Bio Rad RT-PCR RT-PCR 1 icycler iq Bio Rad icycler iq 30 2 Ready-To-Go T-Primed First-Strand Kit (amersham pharmacia biotech) Ready-To-Go T-Primed First-Strand Kit QuantiTect
Real -Time RT-PCR icycler iq Bio Rad RT-PCR RT-PCR 1 icycler iq Bio Rad icycler iq 30 2 Ready-To-Go T-Primed First-Strand Kit (amersham pharmacia biotech) Ready-To-Go T-Primed First-Strand Kit QuantiTect
& Vol.5 No (Oct. 2015) TV 1,2,a) , Augmented TV TV AR Augmented Reality 3DCG TV Estimation of TV Screen Position and Ro
 TV 1,2,a) 1 2 2015 1 26, 2015 5 21 Augmented TV TV AR Augmented Reality 3DCG TV Estimation of TV Screen Position and Rotation Using Mobile Device Hiroyuki Kawakita 1,2,a) Toshio Nakagawa 1 Makoto Sato
TV 1,2,a) 1 2 2015 1 26, 2015 5 21 Augmented TV TV AR Augmented Reality 3DCG TV Estimation of TV Screen Position and Rotation Using Mobile Device Hiroyuki Kawakita 1,2,a) Toshio Nakagawa 1 Makoto Sato
Chapter 1 1-1 2
 Chapter 1 1-1 2 create table ( date, weather ); create table ( date, ); 1 weather, 2 weather, 3 weather, : : 31 weather -- 1 -- 2 -- 3 -- 31 create table ( date, ); weather[] -- 3 Chapter 1 weather[] create
Chapter 1 1-1 2 create table ( date, weather ); create table ( date, ); 1 weather, 2 weather, 3 weather, : : 31 weather -- 1 -- 2 -- 3 -- 31 create table ( date, ); weather[] -- 3 Chapter 1 weather[] create
ScanFront 220/220P 取扱説明書
 libtiff Copyright (c) 1988-1996 Sam Leffler Copyright (c) 1991-1996 Silicon Graphics, Inc. Permission to use, copy, modify, distribute, and sell this software and its documentation for any purpose is hereby
libtiff Copyright (c) 1988-1996 Sam Leffler Copyright (c) 1991-1996 Silicon Graphics, Inc. Permission to use, copy, modify, distribute, and sell this software and its documentation for any purpose is hereby
ScanFront 220/220P セットアップガイド
 libtiff Copyright (c) 1988-1996 Sam Leffler Copyright (c) 1991-1996 Silicon Graphics, Inc. Permission to use, copy, modify, distribute, and sell this software and its documentation for any purpose is hereby
libtiff Copyright (c) 1988-1996 Sam Leffler Copyright (c) 1991-1996 Silicon Graphics, Inc. Permission to use, copy, modify, distribute, and sell this software and its documentation for any purpose is hereby
操作ガイド(本体操作編)
 J-1 QT5-0681-V02 1 m a b c d e f l kj i h g a b c d e f g h i j k l m n n o o s p q r p q r s w t u v x y z t u v w x y z a bc d e f g q p o n m l k j i h a b c d e f g h i j k l {}[] {}[] m n
J-1 QT5-0681-V02 1 m a b c d e f l kj i h g a b c d e f g h i j k l m n n o o s p q r p q r s w t u v x y z t u v w x y z a bc d e f g q p o n m l k j i h a b c d e f g h i j k l {}[] {}[] m n
プレゼンテーション2.ppt
 [email protected] BLAST Genome browser InterProScan PSORT DBTSS Seqlogo JASPAR Melina II Panther Babelomics +@ >cdna_test CCCCTGCCCTCAACAAGATGTTTTGCCAACTGGCCAAGACCTGCCCTGTGCAGCTGTGGGTTGATTCCAC ACCCCCGCCCGGCACCCGCGTCCGCGCCATGGCCATCTACAAGCAGTCACAGCACATGACGGAGGTTGTG
[email protected] BLAST Genome browser InterProScan PSORT DBTSS Seqlogo JASPAR Melina II Panther Babelomics +@ >cdna_test CCCCTGCCCTCAACAAGATGTTTTGCCAACTGGCCAAGACCTGCCCTGTGCAGCTGTGGGTTGATTCCAC ACCCCCGCCCGGCACCCGCGTCCGCGCCATGGCCATCTACAAGCAGTCACAGCACATGACGGAGGTTGTG
GT-X980
 NPD5061-00 JA ...6...10...10...11...13...15...20...21...21...22 /...23 PDF...27 PDF...31 /...35...38...43...46 EPSON Scan...49...49...49...50 EPSON Scan...51...51...52...52...53 2 Windows...53 Mac OS X...53...53...53...54...56...56...58...59...60...60...61...62...63
NPD5061-00 JA ...6...10...10...11...13...15...20...21...21...22 /...23 PDF...27 PDF...31 /...35...38...43...46 EPSON Scan...49...49...49...50 EPSON Scan...51...51...52...52...53 2 Windows...53 Mac OS X...53...53...53...54...56...56...58...59...60...60...61...62...63
GT-X830
 NPD5108-00 ...5... 5... 6... 8...11 EPSON Scan...11 PDF...16 OCR...16...17...17...20 /...20...20...22...23...23...24...25...25...26...27 PDF...30...31 / EPSON Scan...34 EPSON Scan...34 EPSON Scan...36
NPD5108-00 ...5... 5... 6... 8...11 EPSON Scan...11 PDF...16 OCR...16...17...17...20 /...20...20...22...23...23...24...25...25...26...27 PDF...30...31 / EPSON Scan...34 EPSON Scan...34 EPSON Scan...36
PFS-Readme
 Cell Storage Service : Storage Service that has the high reliability and the high availability by Paxos consensus algorithm. CSS-Readme.txt : This manual describes the process to make the envirornments
Cell Storage Service : Storage Service that has the high reliability and the high availability by Paxos consensus algorithm. CSS-Readme.txt : This manual describes the process to make the envirornments
たのしいプログラミング Pythonではじめよう!
 Title of English-language original: Python for Kids A Playful Introduction to Programming ISBN 978-1-59327-407-8, published by No Starch Press, Inc. Copyright 2013 by Jason R. Briggs. Japanese-language
Title of English-language original: Python for Kids A Playful Introduction to Programming ISBN 978-1-59327-407-8, published by No Starch Press, Inc. Copyright 2013 by Jason R. Briggs. Japanese-language
EPSON Easy Interactive Tools Ver.4.0 Operation Guide
 Easy Interactive Tools Ver.4.0 Easy Interactive Tools Ver.4.0 Easy Interactive Tools Easy Interactive Tools Easy Interactive Pen s p.11 s p.10 s p.9 s p.15 s p.17 s p.25 Easy Interactive Tools PowerPoint
Easy Interactive Tools Ver.4.0 Easy Interactive Tools Ver.4.0 Easy Interactive Tools Easy Interactive Tools Easy Interactive Pen s p.11 s p.10 s p.9 s p.15 s p.17 s p.25 Easy Interactive Tools PowerPoint
Actual ESS Adapterの使用について
 Actual ESS Adapter SQL External SQL Source FileMaker SQL ESS SQL FileMaker FileMaker SQL FileMaker FileMaker ESS SQL SQL FileMaker ODBC SQL FileMaker Microsoft SQL Server MySQL Oracle 3 ODBC Mac OS X Actual
Actual ESS Adapter SQL External SQL Source FileMaker SQL ESS SQL FileMaker FileMaker SQL FileMaker FileMaker ESS SQL SQL FileMaker ODBC SQL FileMaker Microsoft SQL Server MySQL Oracle 3 ODBC Mac OS X Actual
TH-47LFX60 / TH-47LFX6N
 TH-47LFX60J TH-47LFX6NJ 1 2 3 4 - + - + DVI-D IN PC IN SERIAL IN AUDIO IN (DVI-D / PC) LAN, DIGITAL LINK AV IN AUDIO OUT 1 11 2 12 3 13 4 14 5 6 15 7 16 8 17 9 18 10 19 19 3 1 18 4 2 HDMI AV OUT
TH-47LFX60J TH-47LFX6NJ 1 2 3 4 - + - + DVI-D IN PC IN SERIAL IN AUDIO IN (DVI-D / PC) LAN, DIGITAL LINK AV IN AUDIO OUT 1 11 2 12 3 13 4 14 5 6 15 7 16 8 17 9 18 10 19 19 3 1 18 4 2 HDMI AV OUT
KNOB Knoppix for Bio Itoshi NIKAIDO
 KNOB Knoppix for Bio Itoshi NIKAIDO Linux Grasp the KNOB! grasp 1, (grip). 2,, (understand). [ 2 ] What s KNOB CD Linux Bioinformatics KNOB Why KNOB? Bioinformatics What
KNOB Knoppix for Bio Itoshi NIKAIDO Linux Grasp the KNOB! grasp 1, (grip). 2,, (understand). [ 2 ] What s KNOB CD Linux Bioinformatics KNOB Why KNOB? Bioinformatics What
How to read the marks and remarks used in this parts book. Section 1 : Explanation of Code Use In MRK Column OO : Interchangeable between the new part
 Reservdelskatalog MIKASA MT65H vibratorstamp EPOX Maskin AB Postadress Besöksadress Telefon Fax e-post Hemsida Version Box 6060 Landsvägen 1 08-754 71 60 08-754 81 00 [email protected] www.epox.se 1,0 192 06
Reservdelskatalog MIKASA MT65H vibratorstamp EPOX Maskin AB Postadress Besöksadress Telefon Fax e-post Hemsida Version Box 6060 Landsvägen 1 08-754 71 60 08-754 81 00 [email protected] www.epox.se 1,0 192 06
ES-D400/ES-D350
 NPD4650-00 ...4 EPSON Scan... 4 Document Capture Pro Windows... 7 EPSON Scan...10 EPSON Scan...10...14 PDF...15 / EPSON Scan...17 EPSON Scan...17 EPSON Scan...18 EPSON Scan...18 Document Capture Pro Windows...19
NPD4650-00 ...4 EPSON Scan... 4 Document Capture Pro Windows... 7 EPSON Scan...10 EPSON Scan...10...14 PDF...15 / EPSON Scan...17 EPSON Scan...17 EPSON Scan...18 EPSON Scan...18 Document Capture Pro Windows...19
Compiled MODELSでのDFT位相検出装置のモデル化と評価
 listsize TPBIG.EXE /Mingw32 ATP (Alternative Transients Program)- EMTP ATP ATP ATP ATP(TPBIG.EXE) EMTP (ATP)FORTAN77 DIMENSION C malloc listsize TACS DIMENSIONEMTP ATP(TPBIG.EXE) listsize (CPU ) RL 4040
listsize TPBIG.EXE /Mingw32 ATP (Alternative Transients Program)- EMTP ATP ATP ATP ATP(TPBIG.EXE) EMTP (ATP)FORTAN77 DIMENSION C malloc listsize TACS DIMENSIONEMTP ATP(TPBIG.EXE) listsize (CPU ) RL 4040
HA8000-bdシリーズ RAID設定ガイド HA8000-bd/BD10X2
 HB102050A0-4 制限 補足 Esc Enter Esc Enter Esc Enter Main Advanced Server Security Boot Exit A SATA Configuration SATA Controller(s) SATA Mode Selection [Enabled] [RAID] Determines how
HB102050A0-4 制限 補足 Esc Enter Esc Enter Esc Enter Main Advanced Server Security Boot Exit A SATA Configuration SATA Controller(s) SATA Mode Selection [Enabled] [RAID] Determines how
How to read the marks and remarks used in this parts book. Section 1 : Explanation of Code Use In MRK Column OO : Interchangeable between the new part
 Reservdelskatalog MIKASA MCD-L14 asfalt- och betongsåg EPOX Maskin AB Postadress Besöksadress Telefon Fax e-post Hemsida Version Box 6060 Landsvägen 1 08-754 71 60 08-754 81 00 [email protected] www.epox.se
Reservdelskatalog MIKASA MCD-L14 asfalt- och betongsåg EPOX Maskin AB Postadress Besöksadress Telefon Fax e-post Hemsida Version Box 6060 Landsvägen 1 08-754 71 60 08-754 81 00 [email protected] www.epox.se
基本操作ガイド
 HT7-0022-000-V.4.0 Copyright 2004 CANON INC. ALL RIGHTS RESERVED 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 4 1 1 2 3 4 5 1 2 1 2 3 1 2 3 1 2 3 1 2 3 4 1 2 3 4 1 2 3 4 5 6 1 2 3 4 5 6 7 1 2 3 4
HT7-0022-000-V.4.0 Copyright 2004 CANON INC. ALL RIGHTS RESERVED 1 2 3 1 2 3 1 2 3 1 2 3 1 2 3 4 1 1 2 3 4 5 1 2 1 2 3 1 2 3 1 2 3 1 2 3 4 1 2 3 4 1 2 3 4 5 6 1 2 3 4 5 6 7 1 2 3 4
1 1.1 (JCPRG) 30 Nuclear Reaction Data File (NRDF) PC GSYS2.4 JCPRG GSYS2.4 Java Windows, Linux, Max OS X, FreeBSD GUI PNG, GIF, JPEG X Y GSYS2
 (GSYS2.4) GSYS2.4 Manual SUZUKI Ryusuke Hokkaido University Hospital Abstract GSYS2.4 is an update version of GSYS version 2. Main features added in this version are Magnifying glass function, Automatically
(GSYS2.4) GSYS2.4 Manual SUZUKI Ryusuke Hokkaido University Hospital Abstract GSYS2.4 is an update version of GSYS version 2. Main features added in this version are Magnifying glass function, Automatically
チュートリアル XP Embedded 入門編
 TUT-0057 Ver. 1.0 www.interface.co.jp Ver 1.0 2005 6 (,), Web site () / () 2004 Interface Corporation. All rights reserved. ...1...1 1. XP Embedded...2 2....3 2.1....3 2.2....4 2.2.1. SLD...4 2.3....5
TUT-0057 Ver. 1.0 www.interface.co.jp Ver 1.0 2005 6 (,), Web site () / () 2004 Interface Corporation. All rights reserved. ...1...1 1. XP Embedded...2 2....3 2.1....3 2.2....4 2.2.1. SLD...4 2.3....5
1 I EViews View Proc Freeze
 EViews 2017 9 6 1 I EViews 4 1 5 2 10 3 13 4 16 4.1 View.......................................... 17 4.2 Proc.......................................... 22 4.3 Freeze & Name....................................
EViews 2017 9 6 1 I EViews 4 1 5 2 10 3 13 4 16 4.1 View.......................................... 17 4.2 Proc.......................................... 22 4.3 Freeze & Name....................................
ProVAL Recent Projects, ProVAL Online 3 Recent Projects ProVAL Online Show Online Content on the Start Page Page 13
 ProVAL Unit System Enable Recording Log Preferred Language Default File Type Default Project Path ProVAL : Unit SystemUse SI Units SI SI USCS Enable Recording Log Language Default File Type Default Project
ProVAL Unit System Enable Recording Log Preferred Language Default File Type Default Project Path ProVAL : Unit SystemUse SI Units SI SI USCS Enable Recording Log Language Default File Type Default Project
Cleaner XL 1.5 クイックインストールガイド
 Autodesk Cleaner XL 1.5 Contents Cleaner XL 1.5 2 1. Cleaner XL 3 2. Cleaner XL 9 3. Cleaner XL 12 4. Cleaner XL 16 5. 32 2 1. Cleaner XL 1. Cleaner XL Cleaner XL Administrators Cleaner XL Windows Media
Autodesk Cleaner XL 1.5 Contents Cleaner XL 1.5 2 1. Cleaner XL 3 2. Cleaner XL 9 3. Cleaner XL 12 4. Cleaner XL 16 5. 32 2 1. Cleaner XL 1. Cleaner XL Cleaner XL Administrators Cleaner XL Windows Media
kubostat2018d p.2 :? bod size x and fertilization f change seed number? : a statistical model for this example? i response variable seed number : { i
 kubostat2018d p.1 I 2018 (d) model selection and [email protected] http://goo.gl/76c4i 2018 06 25 : 2018 06 21 17:45 1 2 3 4 :? AIC : deviance model selection misunderstanding kubostat2018d (http://goo.gl/76c4i)
kubostat2018d p.1 I 2018 (d) model selection and [email protected] http://goo.gl/76c4i 2018 06 25 : 2018 06 21 17:45 1 2 3 4 :? AIC : deviance model selection misunderstanding kubostat2018d (http://goo.gl/76c4i)
GNU Emacs GNU Emacs
 GNU Emacs 2015 10 2 1 GNU Emacs 1 1.1....................................... 1 1.2....................................... 1 1.2.1..................................... 1 1.2.2.....................................
GNU Emacs 2015 10 2 1 GNU Emacs 1 1.1....................................... 1 1.2....................................... 1 1.2.1..................................... 1 1.2.2.....................................
浜松医科大学紀要
 On the Statistical Bias Found in the Horse Racing Data (1) Akio NODA Mathematics Abstract: The purpose of the present paper is to report what type of statistical bias the author has found in the horse
On the Statistical Bias Found in the Horse Racing Data (1) Akio NODA Mathematics Abstract: The purpose of the present paper is to report what type of statistical bias the author has found in the horse
分子系統樹作成方法
 実習 1: MEGA7 のダウンロードとインストール MEGA の Web サイトは http://www.megasoftware.net/( 下図 ) 正式には 黄緑色の [DOWNLOAD] ボタンをクリ ックし 次の画面でもう一度 [DOWNLOAD] をクリックすると ダウンロードできる 実習 2: 配列データのダウンロードとアライメント 例題データ (data): Actin gene
実習 1: MEGA7 のダウンロードとインストール MEGA の Web サイトは http://www.megasoftware.net/( 下図 ) 正式には 黄緑色の [DOWNLOAD] ボタンをクリ ックし 次の画面でもう一度 [DOWNLOAD] をクリックすると ダウンロードできる 実習 2: 配列データのダウンロードとアライメント 例題データ (data): Actin gene
1. MEGA 5 をインストールする 1.1 ダウンロード手順 MEGA のホームページ (http://www.megasoftware.net/index.php) から MEGA 5 software をコンピュータにインストールする 2. 塩基配列を決定する 2.1 Alignment E
 MEGA 5 を用いた塩基配列解析法および分子系統樹作成法 Ver.1 Update: 2012.04.01 ウイルス 疫学研究領域井関博 < 内容 > 1. MEGA 5 をインストールする 1.1 ダウンロード手順 2. 塩基配列を決定する 2.1 Alignment Explorer の起動 2.2 シークエンスデータの入力 2.2.1 テキストファイルから読み込む場合 2.2.2 波形データから読み込む場合
MEGA 5 を用いた塩基配列解析法および分子系統樹作成法 Ver.1 Update: 2012.04.01 ウイルス 疫学研究領域井関博 < 内容 > 1. MEGA 5 をインストールする 1.1 ダウンロード手順 2. 塩基配列を決定する 2.1 Alignment Explorer の起動 2.2 シークエンスデータの入力 2.2.1 テキストファイルから読み込む場合 2.2.2 波形データから読み込む場合
untitled
 SUBJECT: Applied Biosystems Data Collection Software v2.0 v3.0 Windows 2000 OS : 30 45 Cancel Data Collection - Applied Biosystems Sequencing Analysis Software v5.2 - Applied Biosystems SeqScape Software
SUBJECT: Applied Biosystems Data Collection Software v2.0 v3.0 Windows 2000 OS : 30 45 Cancel Data Collection - Applied Biosystems Sequencing Analysis Software v5.2 - Applied Biosystems SeqScape Software
Rによる計量分析:データ解析と可視化 - 第2回 セットアップ
 R 2 2017 Email: [email protected] October 16, 2017 Outline 1 ( ) 2 R RStudio 3 4 R (Toyama/NIHU) R October 16, 2017 1 / 34 R RStudio, R PC ( ) ( ) (Toyama/NIHU) R October 16, 2017 2 / 34 R ( ) R
R 2 2017 Email: [email protected] October 16, 2017 Outline 1 ( ) 2 R RStudio 3 4 R (Toyama/NIHU) R October 16, 2017 1 / 34 R RStudio, R PC ( ) ( ) (Toyama/NIHU) R October 16, 2017 2 / 34 R ( ) R
Zinstall WinWin 日本語ユーザーズガイド
 Zinstall WinWin User Guide Thank you for purchasing Zinstall WinWin. If you have any questions, issues or problems, please contact us: Toll-free phone: (877) 444-1588 International callers: +1-877-444-1588
Zinstall WinWin User Guide Thank you for purchasing Zinstall WinWin. If you have any questions, issues or problems, please contact us: Toll-free phone: (877) 444-1588 International callers: +1-877-444-1588
X Window System X X &
 1 1 1.1 X Window System................................... 1 1.2 X......................................... 1 1.3 X &................................ 1 1.3.1 X.......................... 1 1.3.2 &....................................
1 1 1.1 X Window System................................... 1 1.2 X......................................... 1 1.3 X &................................ 1 1.3.1 X.......................... 1 1.3.2 &....................................
189 2015 1 80
 189 2015 1 A Design and Implementation of the Digital Annotation Basis on an Image Resource for a Touch Operation TSUDA Mitsuhiro 79 189 2015 1 80 81 189 2015 1 82 83 189 2015 1 84 85 189 2015 1 86 87
189 2015 1 A Design and Implementation of the Digital Annotation Basis on an Image Resource for a Touch Operation TSUDA Mitsuhiro 79 189 2015 1 80 81 189 2015 1 82 83 189 2015 1 84 85 189 2015 1 86 87
 WQD770W WQD770W WQD770W WQD770W WQD770W 5 2 1 4 3 WQD8438 WQD770W 1 2 3 5 4 6 7 8 10 12 11 14 13 9 15 16 17 19 20 20 18 21 22 22 24 25 23 2 1 3 1 2 2 3 1 4 1 2 3 2 1 1 2 5 6 3 4 1 2 5 4 6 3 7 8 10 11
WQD770W WQD770W WQD770W WQD770W WQD770W 5 2 1 4 3 WQD8438 WQD770W 1 2 3 5 4 6 7 8 10 12 11 14 13 9 15 16 17 19 20 20 18 21 22 22 24 25 23 2 1 3 1 2 2 3 1 4 1 2 3 2 1 1 2 5 6 3 4 1 2 5 4 6 3 7 8 10 11
NetVehicle GX5取扱説明書 基本編
 -GX5 1 2 3 4 5 6 7 8 # @(#)COPYRIGHT 8.2 (Berkeley) 3/21/94 All of the documentation and software included in the 4.4BSD and 4.4BSD-Lite Releases is copyrighted by The Regents of the University of California.
-GX5 1 2 3 4 5 6 7 8 # @(#)COPYRIGHT 8.2 (Berkeley) 3/21/94 All of the documentation and software included in the 4.4BSD and 4.4BSD-Lite Releases is copyrighted by The Regents of the University of California.
Microsoft Word - PCM TL-Ed.4.4(特定電気用品適合性検査申込のご案内)
 (2017.04 29 36 234 9 1 1. (1) 3 (2) 9 1 2 2. (1) 9 1 1 2 1 2 (2) 1 2 ( PSE-RE-101/205/306/405 2 PSE-RE-201 PSE-RE-301 PSE-RE-401 PSE-RE-302 PSE-RE-202 PSE-RE-303 PSE-RE-402 PSE-RE-203 PSE-RE-304 PSE-RE-403
(2017.04 29 36 234 9 1 1. (1) 3 (2) 9 1 2 2. (1) 9 1 1 2 1 2 (2) 1 2 ( PSE-RE-101/205/306/405 2 PSE-RE-201 PSE-RE-301 PSE-RE-401 PSE-RE-302 PSE-RE-202 PSE-RE-303 PSE-RE-402 PSE-RE-203 PSE-RE-304 PSE-RE-403
nopcommerce 2.2 2.1.6 Adobe Flash ( 1 ) 1 nopcommerce 2.2 ( [5, p.3-4] )
![nopcommerce 2.2 2.1.6 Adobe Flash ( 1 ) 1 nopcommerce 2.2 ( [5, p.3-4] ) nopcommerce 2.2 2.1.6 Adobe Flash ( 1 ) 1 nopcommerce 2.2 ( [5, p.3-4] )](/thumbs/39/20122401.jpg) nopcommerce 2.2 NopCommerce (Ver.2.3) NopCommerce 2.1.1 (OS) Windows 7 Windows Vista Windows XP Windows Server 2003 Windows Server 2008 2.1.2 Web Internet Information Service (IIS) 6.0 2.1.3 ASP.NET 4.0
nopcommerce 2.2 NopCommerce (Ver.2.3) NopCommerce 2.1.1 (OS) Windows 7 Windows Vista Windows XP Windows Server 2003 Windows Server 2008 2.1.2 Web Internet Information Service (IIS) 6.0 2.1.3 ASP.NET 4.0
How to read the marks and remarks used in this parts book. Section 1 : Explanation of Code Use In MRK Column OO : Interchangeable between the new part
 Reservdelskatalog MIKASA MVC-88 vibratorplatta EPOX Maskin AB Postadress Besöksadress Telefon Fax e-post Hemsida Version Box 6060 Landsvägen 1 08-754 71 60 08-754 81 00 [email protected] www.epox.se 1,0 192
Reservdelskatalog MIKASA MVC-88 vibratorplatta EPOX Maskin AB Postadress Besöksadress Telefon Fax e-post Hemsida Version Box 6060 Landsvägen 1 08-754 71 60 08-754 81 00 [email protected] www.epox.se 1,0 192
New version (2.15.1) of Specview is now available Dismiss Windows Specview.bat set spv= Specview set jhome= JAVA (C:\Program Files\Java\jre<version>\
 Specview VO 2012 2012/3/26 Specview Specview STSCI(Space Telescope SCience Institute) VO Specview Web page http://www.stsci.edu/resources/software hardware/specview http://specview.stsci.edu/javahelp/main.html
Specview VO 2012 2012/3/26 Specview Specview STSCI(Space Telescope SCience Institute) VO Specview Web page http://www.stsci.edu/resources/software hardware/specview http://specview.stsci.edu/javahelp/main.html
